Widespread aggregation and neurodegenerative diseases are associated with supersaturated proteins - PubMed (original) (raw)
Widespread aggregation and neurodegenerative diseases are associated with supersaturated proteins
Prajwal Ciryam et al. Cell Rep. 2013.
Abstract
The maintenance of protein solubility is a fundamental aspect of cellular homeostasis because protein aggregation is associated with a wide variety of human diseases. Numerous proteins unrelated in sequence and structure, however, can misfold and aggregate, and widespread aggregation can occur in living systems under stress or aging. A crucial question in this context is why only certain proteins appear to aggregate readily in vivo, whereas others do not. We identify here the proteins most vulnerable to aggregation as those whose cellular concentrations are high relative to their solubilities. We find that these supersaturated proteins represent a metastable subproteome involved in pathological aggregation during stress and aging and are overrepresented in biochemical processes associated with neurodegenerative disorders. Consequently, such cellular processes become dysfunctional when the ability to keep intrinsically supersaturated proteins soluble is compromised. Thus, the simultaneous analysis of abundance and solubility can rationalize the diverse cellular pathologies linked to neurodegenerative diseases and aging.
Copyright © 2013 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
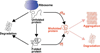
Figure 1. Protein aggregation in vivo can occur through different routes
Proteins may misfold and aggregate as they emerge from the ribosome or when they unfold at least transiently from the native state. The proteins most at risk of aggregation are those whose concentration is high with respect to their solubility. We quantify this risk by defining the supersaturation σ_u_ and σ_f_ scores. The supersaturation score σ_u_, which measures the tendency of proteins to aggregate from the unfolded state, is based on the Zyggregator score (Tartaglia et al., 2008) (Zagg) and mRNA expression levels. The supersaturation score σ_f_, which measures the tendency of proteins to aggregate from the folded state, is based on the structurally-corrected Zyggregator score (Tartaglia et al., 2008) (ZaggSC) and protein concentration.
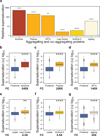
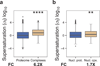
Figure 2. Widespread protein aggregation is associated with high supersaturation scores
(a) Summary of the results for the different classes of aggregating proteins analyzed in this work. Results are given in terms of the increases in the supersaturation scores over the average value for the whole proteome or for an experimental lysate (“fold change”). We compared the supersaturation scores σ_f_ for: (b) the whole proteome and the human “amyloid proteins” in UniProt (UniProt Consortium, 2012), (c) the whole proteome and proteins that co-precipitate in amyloid plaques (Liao et al., 2004), (d) the whole proteome and proteins that co-precipitate in neurofibrillary tangles (Wang et al., 2005), (e) the whole lysate and proteins that co-precipitate in Lewy bodies (Xia et al., 2008), (f) the whole lysate and proteins that co-precipitate in artificial β-sheet protein aggregates (Olzscha et al.), (g) the whole proteome and proteins found to aggregate in C. elegans during aging (David et al., 2010; Reis-Rodrigues et al., 2012). Boxplots extend from the lower to the upper quartiles, with the internal lines refer to the median values. Whiskers range from the lowest to highest value data points within 150% of the interquartile ranges. The statistical significance was assessed by Wilcoxon/Mann-Whitney U test with the Bonferroni-corrected p-values (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
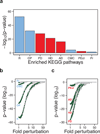
Figure 3. Biochemical processes associated with neurodegenerative diseases are highly enriched in supersaturated proteins
(a) List of the KEGG pathways (Kanehisa et al., 2010) identified here as significantly enriched (Bonferroni-corrected p-values) in proteins at or above the 95th percentile of supersaturation (σ_u_): (R) ribosome, (OP) oxidative phosphorylation, (PD) Parkinson’s disease, (HD) Huntington’s disease, (AD) Alzheimer’s disease, (CMC) cardiac muscle contraction, (PEcI) pathogenic E. coli infection, (Pr) proteasome; physiological and pathological pathways are shown in blue and red, respectively. (b,c) Test of the robustness of the significance of the enrichment of the KEGG pathways according to their supersaturation scores. Gaussian noise was introduced 100 independent times into the proteome scores at 50 different levels and plotted (1× = no noise) for: (b) physiological pathways, which are robust up to 28× (R), 8.8× (OP), 2.3× (CMC) and 1.2× (Pr), and (c) pathological pathways, which are robust up to 8.2× (PD), 6.2× (HD), 5.5× (AD) and 2.1× (PEcI). Error bars indicate interquartile ranges, green points indicate error levels below the p=0.05 significance (red dashed line) by the Wilcoxon/Mann-Whitney U test.
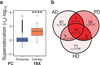
Figure 4. Supersaturated proteins are common to different neurodegenerative pathways
(a) Comparison of the σ_u_ scores for the proteome and the set of 76 proteins common among the Alzheimer’s, Parkinson’s, and Huntington’s KEGG pathways (Kanehisa et al., 2010); this set of proteins is denoted as ‘overlap’. (b) Comparison of the scores for the proteins in the Alzheimer’s, Parkinson’s, and Huntington’s pathways. Colors are assigned based on the division of the σ_u_ scores into deciles from low (green) to high (red).
Figure 5. Proteins that form complexes tend to be more supersaturated than the proteome as a whole
We compared the σ_f_ scores for: (a) the proteome and proteins involved in complexes, and (b) the nuclear proteome and proteins involved in nuclear complexes. Boxplots extend from the lower to the upper quartiles, with the internal lines refer to the median values. Whiskers range from the lowest to highest value data points within 150% of the interquartile ranges. The statistical significance was assessed by Wilcoxon/Mann-Whitney U test with the Bonferroni-corrected p-values (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
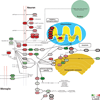
Figure 6. Supersaturated proteins are over-represented in the KEGG Alzheimer’s disease pathway
The KEGG pathway for Alzheimer’s disease (Kanehisa et al.) is curated from the literature and includes many proteins not directly associated with the disruption of the homeostasis of the Aβ peptide. Those proteins that are part of the overlap (see Fig. 4a) of the Alzheimer’s, Parkinson’s, and Huntington’s disease pathways are shown in bold. Approximately 65% of proteins in the Alzheimer’s pathway have high σ_u_ scores. Colors are assigned based on the division of the σ_u_ database into deciles from low (green) to high (red).
Similar articles
- Supersaturation is a major driving force for protein aggregation in neurodegenerative diseases.
Ciryam P, Kundra R, Morimoto RI, Dobson CM, Vendruscolo M. Ciryam P, et al. Trends Pharmacol Sci. 2015 Feb;36(2):72-7. doi: 10.1016/j.tips.2014.12.004. Epub 2015 Jan 27. Trends Pharmacol Sci. 2015. PMID: 25636813 Free PMC article. Review. - Role of metal ions in aggregation of intrinsically disordered proteins in neurodegenerative diseases.
Breydo L, Uversky VN. Breydo L, et al. Metallomics. 2011 Nov;3(11):1163-80. doi: 10.1039/c1mt00106j. Epub 2011 Aug 25. Metallomics. 2011. PMID: 21869995 Review. - A metastable subproteome underlies inclusion formation in muscle proteinopathies.
Ciryam P, Antalek M, Cid F, Tartaglia GG, Dobson CM, Guettsches AK, Eggers B, Vorgerd M, Marcus K, Kley RA, Morimoto RI, Vendruscolo M, Weihl CC. Ciryam P, et al. Acta Neuropathol Commun. 2019 Dec 3;7(1):197. doi: 10.1186/s40478-019-0853-9. Acta Neuropathol Commun. 2019. PMID: 31796104 Free PMC article. - Defective protein folding and aggregation as the basis of neurodegenerative diseases: the darker aspect of proteins.
Naeem A, Fazili NA. Naeem A, et al. Cell Biochem Biophys. 2011 Nov;61(2):237-50. doi: 10.1007/s12013-011-9200-x. Cell Biochem Biophys. 2011. PMID: 21573992 Review. - Targeting DNA topoisomerases or checkpoint kinases results in an overload of chaperone systems, triggering aggregation of a metastable subproteome.
Huiting W, Dekker SL, van der Lienden JCJ, Mergener R, Musskopf MK, Furtado GV, Gerrits E, Coit D, Oghbaie M, Di Stefano LH, Schepers H, van Waarde-Verhagen MAWH, Couzijn S, Barazzuol L, LaCava J, Kampinga HH, Bergink S. Huiting W, et al. Elife. 2022 Feb 24;11:e70726. doi: 10.7554/eLife.70726. Elife. 2022. PMID: 35200138 Free PMC article.
Cited by
- The supersaturation perspective on the amyloid hypothesis.
Portugal Barron D, Guo Z. Portugal Barron D, et al. Chem Sci. 2023 Oct 16;15(1):46-54. doi: 10.1039/d3sc03981a. eCollection 2023 Dec 20. Chem Sci. 2023. PMID: 38131088 Free PMC article. Review. - Proteomics revealed an association between ribosome-associated proteins and amyloid beta deposition in Alzheimer's disease.
Feng L, Wang G, Song Q, Feng X, Su J, Ji G, Li M. Feng L, et al. Metab Brain Dis. 2024 Feb;39(2):263-282. doi: 10.1007/s11011-023-01330-3. Epub 2023 Nov 29. Metab Brain Dis. 2024. PMID: 38019374 - Influence of Cortisol on the Fibril Formation Kinetics of Aβ42 Peptide: A Multi-Technical Approach.
Nucara A, Ripanti F, Sennato S, Nisini G, De Santis E, Sefat M, Carbonaro M, Mango D, Minicozzi V, Carbone M. Nucara A, et al. Int J Mol Sci. 2022 May 26;23(11):6007. doi: 10.3390/ijms23116007. Int J Mol Sci. 2022. PMID: 35682687 Free PMC article. - Impact of Deleterious Mutations on Structure, Function and Stability of Serum/Glucocorticoid Regulated Kinase 1: A Gene to Diseases Correlation.
AlAjmi MF, Khan S, Choudhury A, Mohammad T, Noor S, Hussain A, Lu W, Eapen MS, Chimankar V, Hansbro PM, Sohal SS, Elasbali AM, Hassan MI. AlAjmi MF, et al. Front Mol Biosci. 2021 Nov 3;8:780284. doi: 10.3389/fmolb.2021.780284. eCollection 2021. Front Mol Biosci. 2021. PMID: 34805284 Free PMC article. - Extensive protein dosage compensation in aneuploid human cancers.
Schukken KM, Sheltzer JM. Schukken KM, et al. Genome Res. 2022 Jul;32(7):1254-1270. doi: 10.1101/gr.276378.121. Epub 2022 Jun 14. Genome Res. 2022. PMID: 35701073 Free PMC article.
References
- Agostini F, Vendruscolo M, Tartaglia GG. Sequence-based prediction of protein solubility. J. Mol. Biol. 2012;421:237–241. - PubMed
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. - PubMed
- Baldwin AJ, Knowles TPJ, Tartaglia GG, Fitzpatrick AW, Devlin GL, Shammas SL, Waudby CA, Mossuto MF, Meehan S, Gras SL, et al. Metastability of native proteins and the phenomenon of amyloid formation. J. Am. Chem. Soc. 2011;133:14160–14163. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 GM038109/GM/NIGMS NIH HHS/United States
- R37 AG026647/AG/NIA NIH HHS/United States
- 089703/WT_/Wellcome Trust/United Kingdom
- R01 GM038109/GM/NIGMS NIH HHS/United States
- BB/H003843/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R01 AG026647/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical