PIWI proteins are essential for early Drosophila embryogenesis - PubMed (original) (raw)
PIWI proteins are essential for early Drosophila embryogenesis
Sneha Ramesh Mani et al. Dev Biol. 2014.
Abstract
PIWI proteins, a subfamily of the ARGONAUTE/PIWI protein family, have been implicated in transcriptional and posttranscriptional gene regulation and transposon silencing mediated by small non-coding RNAs, especially piRNAs. Although these proteins are known to be required for germline development, their somatic function remains elusive. Here, we examine the maternal function of all three PIWI proteins in Drosophila; Piwi, Aubergine (Aub) and Argonaute3 (Ago3) during early embryogenesis. In syncytial embryos, Piwi displays an embryonic stage-dependent localization pattern. Piwi is localized in the cytoplasm during mitotic cycles 1-10. Between cycles 11 and 14, Piwi remains in the cytoplasm during mitosis but moves into the somatic nucleus during interphase. Beyond cycle 14, it stays in the nucleus. Aub and Ago3 are diffusely cytoplasmic from cycle 1 to 14. Embryos maternally depleted of any one of the three PIWI proteins display severe mitotic defects, including abnormal chromosome and nuclear morphology, cell cycle arrest, asynchronous nuclear division and aberrant nuclear migration. Furthermore, all three PIWI proteins are required for the assembly of mitotic machinery and progression through mitosis. Embryos depleted of maternal PIWI proteins also exhibit chromatin organization abnormalities. These observations indicate that maternal Piwi, Aub and Ago3 play a critical role in the maintenance of chromatin structure and cell cycle progression during early embryogenesis, with compromised chromatin integrity as a possible cause of the observed mitotic defects. Our study demonstrates the essential function of PIWI proteins in the first phase of somatic development.
Keywords: Argonaute 3; Aubergine; Chromosome; Embryonic; Maternal; Mitosis; PIWI.
© 2013 Elsevier Inc. All rights reserved.
Figures
Figure 1. Piwi exhibits a dynamic localization during early embryogenesis while Aub and Ago3 are cytoplasmic
Confocal micrographs of wildtype embryos stained for DAPI (A-L) Piwi (A’, D’, G’, J’), Aub (B’, E’, H’, K’) or Ago3 (C’, F’, I’, L’). Scale bars on the leftmost image denote scale bars for the whole row. Between cycles 1-8, embryos reveal diffuse cytoplasmic staining for Piwi, Aub and Ago3(A-C’). At cycle 13, Piwi is the nuclei in both somatic pole cells at interphase (D, D’) but is localized to the cytoplasm of somatic nuclei at prophase (G, G’). It remains nuclear in both somatic and pole cells at cycle 14 (J, J’). At cycle 13, Aub remains cytoplasmic at early prophase (E, E’) and metaphase (H,H’) in both somatic embryo and pole cells, and remains so at cycle 14 after the embryo is cellularized (K, K’). Ago3 shows similar cytpolasmic localization in both somatic and pole cell nuclei as Aub from cycle 13 (F, F’, I, I’) to cycle 14 (L, L’). Scale bars on the leftmost image denote scale bars for the whole row.
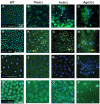
Figure 2. Maternally PIWI-depleted embryos display various mitotic defects
Confocal micrographs of DAPI-stained Piwi-depleted (A,D,G,J,M,P), Aub-depleted(E,H,K,N,Q) and Ago3-depleted (F,I,L,O,R) embryos displaying: (A) Arrest at one cell stage (B,C) Stage of arrest distribution (D-F) Abnormal nuclear morphology, (G-I) Asynchronous nuclear division, (J-L) Abnormal nuclear migration, (M-O) Polar body defects and (P-R) No mitotic defects. Scale bars on the leftmost image denote scale bars for the whole row.
Figure 3. Maternally PIWI-depleted embryos exhibit mitotic abnormalities
Confocal micrographs of wildtype (A, E, I), Piwi- (B, F, J), Aub- (C, G, K) and Ago3- deficient (D, H, L) embryos. (A) Wildtype embryos show normal organization of the spindle apparatus. DAPI is in blue and alpha tubulin is in green. (B-D) Alpha tubulin (green) is stretched and spindle morphology is abnormal in mutants. (E) Two centrosomes per nucleus (closed circle) is seen in wildtype embryos. Centrosomes are highlighted by Centrosomin (Cnn), in red. (F-H) Abnormal Cnn (red) expression is seen in mutant embryos. Dashed circles represent nuclei with too few or too many Cnn positive signals and arrows highlight nuclei with no Cnn signal. (I) All nuclei in wildtype embryos concurrently stain positively for phosphorylated histone 3 (red) (J-L) Nuclei in mutant embryos show variability in PH3 staining. Some nuclei are positive for PH3 while neighboring nuclei are negative (arrowheads). Very fragmented nuclei are also able to enter mitosis (arrows). The scale bar in A represents 10μm for A-L.
Figure 4. Mitotic defects are present during very early nuclear divisions in Aub- and Ago3- deficient embryos
Confocal micrographs of DAPI-stained embryos 30 mins after egg laying (A-E) Wildtype embryos in each stage of mitotic cycle 1 and 2 are easily discernable (F-J) Early Aub-deficient embryos display severe defects including chromosome fragmentation, chromatin bridge formation and condensation abnormalities (K-O) Nuclei in very early Ago3-depleted embryos show segregation defects, chromosome fragmentation and inappropriate condensation. The scale bar in A represents 10μm for A-O.
Figure 5. Maternally PIWI-depleted embryos exhibit defects in nuclear and chromatin organization
Confocal micrographs of wildtype (A,E,I,M) , Piwi-depleted (B,F,J,N), Aub-depleted (C,G,K,O) and Ago3-depleted (D,H,J,P) embryos. (A) Lamin expression in wildtype embryos is even and circular (B-D) Lamin staining in mutant nuclei is highly abnormal and indicates nuclear morphology disruption (E) Even HP1a expression is seen on chromosomes (F-H) Abnormally high HP1a expression (dashed circles) and highly diffuse signal (arrows) are both present in the nuclei examined. (I) Wild type expression of histone 3 Me3K9, a chromatin modification involved in HP1a localization (J) Piwi mutants show a reduction in histone 3 Me3K9 expression (K,L) Histone 3 Me3K9 is below the threshold of detection in Aub and Ago3 mutants (M) Staining against ORC2 in wildtype embryos shows a pattern similar to HP1a (N-P) ORC2 staining in PIWI-deficient embryos is diffuse and excluded from chromatin. The scale bar in A represents 10μm for A-L. The scale bar in (M) represents 10μm in M-P.
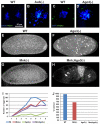
Figure 6. Defects in PIWI-deficient embryos are independent the DNA damage pathway
(A,C) Nuclei in early wildtype embryos show no phosphorylated H2Av (γH2Av) signal (green) (B,D) Upregulation of γH2Av signal is seen in fragmented nuclei of very early Aub- and Ago3- deficient embryos respectively. The scale bar in (A) represents 10μm for A-D. (E) Confocal image of DAPI stained wildtype embryo after 2-3 hours of development. (F) DAPI stained image of age-matched Ago3-depleted embryo shows highly abnormal nuclei. (G) Mnk(-) mutant embryos show normal early development. (H) Embryos laid by Mnk;Ago3 double mutants are phenotypically similar to those laid by Ago3(−) females. (I) The graph shows the number of eggs laid (y axis) as a function of time (x axis) by females of the indicated genotypes. (J) The graph shows percentage of eggs laid that hatch (y axis) for each indicated genotype (x axis)
Similar articles
- Multiple roles for Piwi in silencing Drosophila transposons.
Rozhkov NV, Hammell M, Hannon GJ. Rozhkov NV, et al. Genes Dev. 2013 Feb 15;27(4):400-12. doi: 10.1101/gad.209767.112. Epub 2013 Feb 7. Genes Dev. 2013. PMID: 23392609 Free PMC article. - [The interplay of transposon silencing genes in the Drosophila melanogaster germline].
Sokolova OA, Iakushev EIu, Stoliarenko AD, Mikhaleva EA, Gvozdev VA, Klenov MS. Sokolova OA, et al. Mol Biol (Mosk). 2011 Jul-Aug;45(4):633-41. Mol Biol (Mosk). 2011. PMID: 21954595 Russian. - Slicing and Binding by Ago3 or Aub Trigger Piwi-Bound piRNA Production by Distinct Mechanisms.
Wang W, Han BW, Tipping C, Ge DT, Zhang Z, Weng Z, Zamore PD. Wang W, et al. Mol Cell. 2015 Sep 3;59(5):819-30. doi: 10.1016/j.molcel.2015.08.007. Mol Cell. 2015. PMID: 26340424 Free PMC article. - piRNA-mediated silencing in Drosophila germlines.
Siomi MC, Miyoshi T, Siomi H. Siomi MC, et al. Semin Cell Dev Biol. 2010 Sep;21(7):754-9. doi: 10.1016/j.semcdb.2010.01.011. Epub 2010 Jan 18. Semin Cell Dev Biol. 2010. PMID: 20080197 Review. - Untangling the web: the diverse functions of the PIWI/piRNA pathway.
Mani SR, Juliano CE. Mani SR, et al. Mol Reprod Dev. 2013 Aug;80(8):632-64. doi: 10.1002/mrd.22195. Epub 2013 Jun 27. Mol Reprod Dev. 2013. PMID: 23712694 Free PMC article. Review.
Cited by
- Maternal Proteins That Are Phosphoregulated upon Egg Activation Include Crucial Factors for Oogenesis, Egg Activation and Embryogenesis in Drosophila melanogaster.
Zhang Z, Krauchunas AR, Huang S, Wolfner MF. Zhang Z, et al. G3 (Bethesda). 2018 Aug 30;8(9):3005-3018. doi: 10.1534/g3.118.200578. G3 (Bethesda). 2018. PMID: 30012668 Free PMC article. - GCNA Preserves Genome Integrity and Fertility Across Species.
Bhargava V, Goldstein CD, Russell L, Xu L, Ahmed M, Li W, Casey A, Servage K, Kollipara R, Picciarelli Z, Kittler R, Yatsenko A, Carmell M, Orth K, Amatruda JF, Yanowitz JL, Buszczak M. Bhargava V, et al. Dev Cell. 2020 Jan 6;52(1):38-52.e10. doi: 10.1016/j.devcel.2019.11.007. Epub 2019 Dec 12. Dev Cell. 2020. PMID: 31839537 Free PMC article. - PIWI proteins and their interactors in piRNA biogenesis, germline development and gene expression.
Ku HY, Lin H. Ku HY, et al. Natl Sci Rev. 2014 Jun;1(2):205-218. doi: 10.1093/nsr/nwu014. Natl Sci Rev. 2014. PMID: 25512877 Free PMC article. - Non-Coding RNAs in Glioma.
Rynkeviciene R, Simiene J, Strainiene E, Stankevicius V, Usinskiene J, Miseikyte Kaubriene E, Meskinyte I, Cicenas J, Suziedelis K. Rynkeviciene R, et al. Cancers (Basel). 2018 Dec 22;11(1):17. doi: 10.3390/cancers11010017. Cancers (Basel). 2018. PMID: 30583549 Free PMC article. Review. - Dynamic RNA-protein interactions underlie the zebrafish maternal-to-zygotic transition.
Despic V, Dejung M, Gu M, Krishnan J, Zhang J, Herzel L, Straube K, Gerstein MB, Butter F, Neugebauer KM. Despic V, et al. Genome Res. 2017 Jul;27(7):1184-1194. doi: 10.1101/gr.215954.116. Epub 2017 Apr 5. Genome Res. 2017. PMID: 28381614 Free PMC article.
References
- Abdu U, Brodsky M, Schüpbach T. Activation of a Meiotic Checkpoint during Drosophila Oogenesis Regulates the Translation of Gurken through Chk2/Mnk. Current Biology. 200212:1645–1651. - PubMed
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001410:120–124. - PubMed
- Bartek J, Lukas J. DNA damage checkpoints: from initiation to recovery or adaptation. Current Opinion in Cell Biology. 200719:238–245. - PubMed
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007128:1089–1103. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases