Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas - PubMed (original) (raw)
. 2013 Dec;45(12):1470-1473.
doi: 10.1038/ng.2813. Epub 2013 Nov 3.
Yuchen Jiao # 1 2 3, Timothy M Pawlik # 3 4, Robert A Anders # 3 5, Florin M Selaru 6, Mirte M Streppel 5, Donald J Lucas 7, Noushin Niknafs 8, Violeta Beleva Guthrie 8, Anirban Maitra 3 5, Pedram Argani 3 5, G Johan A Offerhaus 9, Juan Carlos Roa 10, Lewis R Roberts 11, Gregory J Gores 11, Irinel Popescu 12, Sorin T Alexandrescu 12, Simona Dima 12, Matteo Fassan 13 14, Michele Simbolo 13 14, Andrea Mafficini 13, Paola Capelli 14, Rita T Lawlor 13 14, Andrea Ruzzenente 15, Alfredo Guglielmi 15, Giampaolo Tortora 16, Filippo de Braud 17, Aldo Scarpa 13 14, William Jarnagin 18, David Klimstra 19, Rachel Karchin 8, Victor E Velculescu 1 2 3, Ralph H Hruban 3 5, Bert Vogelstein 1 2 3, Kenneth W Kinzler 1 2 3, Nickolas Papadopoulos 1 2 3, Laura D Wood 5
Affiliations
- PMID: 24185509
- PMCID: PMC4013720
- DOI: 10.1038/ng.2813
Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas
Yuchen Jiao et al. Nat Genet. 2013 Dec.
Abstract
Through exomic sequencing of 32 intrahepatic cholangiocarcinomas, we discovered frequent inactivating mutations in multiple chromatin-remodeling genes (including BAP1, ARID1A and PBRM1), and mutation in one of these genes occurred in almost half of the carcinomas sequenced. We also identified frequent mutations at previously reported hotspots in the IDH1 and IDH2 genes encoding metabolic enzymes in intrahepatic cholangiocarcinomas. In contrast, TP53 was the most frequently altered gene in a series of nine gallbladder carcinomas. These discoveries highlight the key role of dysregulated chromatin remodeling in intrahepatic cholangiocarcinomas.
Figures
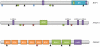
Figure 1
Genes with frequent inactivating mutations in intrahepatic cholangiocarcinoma. Inactivating mutations occurred throughout the coding sequences of BAP1, ARID1A and PBRM1. Rectangles, splice-site mutations; triangles, insertions and deletions; ovals, missense mutations; stars, nonsense mutations. H, HBM-like motif; BR, BRCA1-interacting domain; C, coiled-coil domain; N, nuclear localization signal; LXXLL, LXXLL domain; ARID, ARID domain; BD, bromodomain; BAH, BAH domain; HMG, HMG box.
Comment in
- Genetics: genetics of biliary tract cancer.
Ray K. Ray K. Nat Rev Gastroenterol Hepatol. 2013 Dec;10(12):692. doi: 10.1038/nrgastro.2013.219. Epub 2013 Nov 19. Nat Rev Gastroenterol Hepatol. 2013. PMID: 24247265 No abstract available.
References
- Everhart JE, Ruhl CE. Burden of digestive diseases in the United States Part III: Liver, biliary tract, and pancreas. Gastroenterology. 2009;136:1134–1144. - PubMed
- Voss JS, et al. Molecular profiling of cholangiocarcinoma shows potential for targeted therapy treatment decisions. Hum. Pathol. 2013;44:1216–1222. - PubMed
- Xu RF, et al. KRAS and PIK3CA but not BRAF genes are frequently mutated in Chinese cholangiocarcinoma patients. Biomed. Pharmacother. 2011;65:22–26. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 CA043460/CA/NCI NIH HHS/United States
- EDRN U01CA086402/CA/NCI NIH HHS/United States
- R01 CA121113/CA/NCI NIH HHS/United States
- P50 CA62924/CA/NCI NIH HHS/United States
- K08 DK090154/DK/NIDDK NIH HHS/United States
- P30 CA015083/CA/NCI NIH HHS/United States
- K08DK090154/DK/NIDDK NIH HHS/United States
- P50 CA062924/CA/NCI NIH HHS/United States
- P30 CA006973/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- U01 CA086402/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous