Binding to E1 and E3 is mutually exclusive for the human autophagy E2 Atg3 - PubMed (original) (raw)
Binding to E1 and E3 is mutually exclusive for the human autophagy E2 Atg3
Yu Qiu et al. Protein Sci. 2013 Dec.
Abstract
Ubiquitin-like proteins (UBLs) are activated, transferred and conjugated by E1-E2-E3 enzyme cascades. E2 enzymes for canonical UBLs such as ubiquitin, SUMO, and NEDD8 typically use common surfaces to bind to E1 and E3 enzymes. Thus, canonical E2s are required to disengage from E1 prior to E3-mediated UBL ligation. However, E1, E2, and E3 enzymes in the autophagy pathway are structurally and functionally distinct from canonical enzymes, and it has not been possible to predict whether autophagy UBL cascades are organized according to the same principles. Here, we address this question for the pathway mediating lipidation of the human autophagy UBL, LC3. We utilized bioinformatic and experimental approaches to identify a distinctive region in the autophagy E2, Atg3, that binds to the autophagy E3, Atg12∼Atg5-Atg16. Short peptides corresponding to this Atg3 sequence inhibit LC3 lipidation in vitro. Notably, the E3-binding site on Atg3 overlaps with the binding site for the E1, Atg7. Accordingly, the E3 competes with Atg7 for binding to Atg3, implying that Atg3 likely cycles back and forth between binding to Atg7 for loading with the UBL LC3 and binding to E3 to promote LC3 lipidation. The results show that common organizational principles underlie canonical and noncanonical UBL transfer cascades, but are established through distinct structural features.
Keywords: Atg12∼Atg5; Atg3; Atg7; E1 enzyme; E1‐E2 binding; E2 enzyme; E2‐E3 binding; E3 enzyme; autophagy; ubiquitin‐like protein.
© 2013 The Protein Society.
Figures
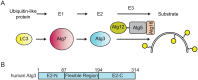
Figure 1
LC3 conjugation pathway. (A) Atg8 family members such as human LC3 are activated by the E1 Atg7 and then transferred to the E2 enzyme, Atg3. Atg12∼Atg5-Atg16 acts as an E3 enzyme that catalyzes lipid conjugation of LC3 and other Atg8 protein family members. (B) Domain diagram representing the primary structure of Atg3 and highlighting the Atg3 Flexible Region. E2-N and E2-C are the N- and C-terminal parts of an E2 fold.
Figure 2
Atg7 and Atg12∼Atg5-Atg16L1 bind to an overlapping region in the Atg3 flexible region (FR). (A) Coomassie-stained SDS-PAGE gels of affinity pulldowns with purified recombinant GST-Atg7 and wild-type (wt) or deletion mutant versions of Atg3. Lanes showing input material are labeled “i” and lanes with affinity pulldowns are labeled “p.” (B) Affinity pulldown with purified recombinant GST-Atg12∼Atg5-his-MBP-Atg16L1 complex and wild-type or deletion mutant versions of Atg3.
Figure 3
Competition experiments. (A,B) 20 μ_M_ Atg7-Atg3 or Atg7-Atg3(Δ145–160) complex was incubated with increasing amounts of Atg12∼Atg5-Atg16L1 complex (0, 4, 10, 20 μ_M_) as described under “Materials and Methods.” Atg3, Atg12∼Atg5-Atg16L1 complex, the Atg7-Atg3 complex, and the Atg3-Atg12∼Atg5-Atg16L1 complex were identified by their different migrations on a Coomassie-stained nondenaturing polyacrylamide gel and are indicated at the right. (C) Binding competition assay performed as for A and B showing titration of Atg3-Atg12∼Atg5-Atg16L1 with Atg7. (D) LC3 lipidation competition assays. Coomassie-stained urea SDS-PAGE showing in vitro LC3 lipidation in multiple turnover assays containing Atg7, Atg3, Atg12∼Atg5, and LC3. GST or GST-Atg3 peptides were added at a 10-fold molar excess over Atg3. (E) LC3 lipidation competition assays performed in the absence of E3 enzyme. Sypro-stained urea SDS-PAGE showing in vitro LC3 lipidation in multiple turnover assays containing Atg7, Atg3, and LC3. GST or GST-Atg3 peptides were added at a 10-fold molar excess over Atg3.
Similar articles
- A switch element in the autophagy E2 Atg3 mediates allosteric regulation across the lipidation cascade.
Zheng Y, Qiu Y, Grace CRR, Liu X, Klionsky DJ, Schulman BA. Zheng Y, et al. Nat Commun. 2019 Aug 9;10(1):3600. doi: 10.1038/s41467-019-11435-y. Nat Commun. 2019. PMID: 31399562 Free PMC article. - Allosteric regulation through a switch element in the autophagy E2, Atg3.
Qiu Y, Zheng Y, Grace CRR, Liu X, Klionsky DJ, Schulman BA. Qiu Y, et al. Autophagy. 2020 Jan;16(1):183-184. doi: 10.1080/15548627.2019.1688550. Epub 2019 Nov 9. Autophagy. 2020. PMID: 31690182 Free PMC article. Review. - Structures of Atg7-Atg3 and Atg7-Atg10 reveal noncanonical mechanisms of E2 recruitment by the autophagy E1.
Kaiser SE, Qiu Y, Coats JE, Mao K, Klionsky DJ, Schulman BA. Kaiser SE, et al. Autophagy. 2013 May;9(5):778-80. doi: 10.4161/auto.23644. Epub 2013 Feb 6. Autophagy. 2013. PMID: 23388412 Free PMC article. - Noncanonical E2 recruitment by the autophagy E1 revealed by Atg7-Atg3 and Atg7-Atg10 structures.
Kaiser SE, Mao K, Taherbhoy AM, Yu S, Olszewski JL, Duda DM, Kurinov I, Deng A, Fenn TD, Klionsky DJ, Schulman BA. Kaiser SE, et al. Nat Struct Mol Biol. 2012 Dec;19(12):1242-9. doi: 10.1038/nsmb.2415. Epub 2012 Nov 11. Nat Struct Mol Biol. 2012. PMID: 23142976 Free PMC article. - Structural insights into E2-E3 interaction for LC3 lipidation.
Metlagel Z, Otomo C, Ohashi K, Takaesu G, Otomo T. Metlagel Z, et al. Autophagy. 2014 Mar;10(3):522-3. doi: 10.4161/auto.27594. Epub 2014 Jan 9. Autophagy. 2014. PMID: 24413923 Free PMC article. Review.
Cited by
- Ubiquitin-like Protein Conjugation: Structures, Chemistry, and Mechanism.
Cappadocia L, Lima CD. Cappadocia L, et al. Chem Rev. 2018 Feb 14;118(3):889-918. doi: 10.1021/acs.chemrev.6b00737. Epub 2017 Feb 24. Chem Rev. 2018. PMID: 28234446 Free PMC article. Review. - Identification and characterization of the linear region of ATG3 that interacts with ATG7 in higher eukaryotes.
Ohashi K, Otomo T. Ohashi K, et al. Biochem Biophys Res Commun. 2015 Jul 31;463(3):447-52. doi: 10.1016/j.bbrc.2015.05.107. Epub 2015 Jun 2. Biochem Biophys Res Commun. 2015. PMID: 26043688 Free PMC article. - Mechanisms and Pathophysiological Roles of the ATG8 Conjugation Machinery.
Lystad AH, Simonsen A. Lystad AH, et al. Cells. 2019 Aug 25;8(9):973. doi: 10.3390/cells8090973. Cells. 2019. PMID: 31450711 Free PMC article. Review. - A switch element in the autophagy E2 Atg3 mediates allosteric regulation across the lipidation cascade.
Zheng Y, Qiu Y, Grace CRR, Liu X, Klionsky DJ, Schulman BA. Zheng Y, et al. Nat Commun. 2019 Aug 9;10(1):3600. doi: 10.1038/s41467-019-11435-y. Nat Commun. 2019. PMID: 31399562 Free PMC article. - Production of Human ATG Proteins for Lipidation Assays.
Zheng Y, Qiu Y, Gunderson JE, Schulman BA. Zheng Y, et al. Methods Enzymol. 2017;587:97-113. doi: 10.1016/bs.mie.2016.09.055. Epub 2016 Nov 11. Methods Enzymol. 2017. PMID: 28253979 Free PMC article.
References
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. - PubMed
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. - PubMed
- Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 5P30CA021765/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 GM077053/GM/NIGMS NIH HHS/United States
- R01GM077053/GM/NIGMS NIH HHS/United States
- P30 CA021765/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous