JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles - PubMed (original) (raw)
. 2014 Jan;42(Database issue):D142-7.
doi: 10.1093/nar/gkt997. Epub 2013 Nov 4.
Xiaobei Zhao, Allen W Zhang, François Parcy, Rebecca Worsley-Hunt, David J Arenillas, Sorana Buchman, Chih-yu Chen, Alice Chou, Hans Ienasescu, Jonathan Lim, Casper Shyr, Ge Tan, Michelle Zhou, Boris Lenhard, Albin Sandelin, Wyeth W Wasserman
Affiliations
- PMID: 24194598
- PMCID: PMC3965086
- DOI: 10.1093/nar/gkt997
JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles
Anthony Mathelier et al. Nucleic Acids Res. 2014 Jan.
Abstract
JASPAR (http://jaspar.genereg.net) is the largest open-access database of matrix-based nucleotide profiles describing the binding preference of transcription factors from multiple species. The fifth major release greatly expands the heart of JASPAR-the JASPAR CORE subcollection, which contains curated, non-redundant profiles-with 135 new curated profiles (74 in vertebrates, 8 in Drosophila melanogaster, 10 in Caenorhabditis elegans and 43 in Arabidopsis thaliana; a 30% increase in total) and 43 older updated profiles (36 in vertebrates, 3 in D. melanogaster and 4 in A. thaliana; a 9% update in total). The new and updated profiles are mainly derived from published chromatin immunoprecipitation-seq experimental datasets. In addition, the web interface has been enhanced with advanced capabilities in browsing, searching and subsetting. Finally, the new JASPAR release is accompanied by a new BioPython package, a new R tool package and a new R/Bioconductor data package to facilitate access for both manual and automated methods.
Figures
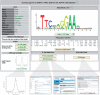
Figure 1.
Screenshot of an example TFBS profile in new layout.
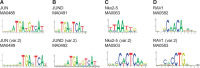
Figure 2.
TFBSs with two different profiles. (A) JUN, (B) JUND, (C) Nkx2-5 and (D) RAV1.
Similar articles
- JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles.
Mathelier A, Fornes O, Arenillas DJ, Chen CY, Denay G, Lee J, Shi W, Shyr C, Tan G, Worsley-Hunt R, Zhang AW, Parcy F, Lenhard B, Sandelin A, Wasserman WW. Mathelier A, et al. Nucleic Acids Res. 2016 Jan 4;44(D1):D110-5. doi: 10.1093/nar/gkv1176. Epub 2015 Nov 3. Nucleic Acids Res. 2016. PMID: 26531826 Free PMC article. - JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework.
Khan A, Fornes O, Stigliani A, Gheorghe M, Castro-Mondragon JA, van der Lee R, Bessy A, Chèneby J, Kulkarni SR, Tan G, Baranasic D, Arenillas DJ, Sandelin A, Vandepoele K, Lenhard B, Ballester B, Wasserman WW, Parcy F, Mathelier A. Khan A, et al. Nucleic Acids Res. 2018 Jan 4;46(D1):D260-D266. doi: 10.1093/nar/gkx1126. Nucleic Acids Res. 2018. PMID: 29140473 Free PMC article. - JASPAR 2020: update of the open-access database of transcription factor binding profiles.
Fornes O, Castro-Mondragon JA, Khan A, van der Lee R, Zhang X, Richmond PA, Modi BP, Correard S, Gheorghe M, Baranašić D, Santana-Garcia W, Tan G, Chèneby J, Ballester B, Parcy F, Sandelin A, Lenhard B, Wasserman WW, Mathelier A. Fornes O, et al. Nucleic Acids Res. 2020 Jan 8;48(D1):D87-D92. doi: 10.1093/nar/gkz1001. Nucleic Acids Res. 2020. PMID: 31701148 Free PMC article. - JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update.
Bryne JC, Valen E, Tang MH, Marstrand T, Winther O, da Piedade I, Krogh A, Lenhard B, Sandelin A. Bryne JC, et al. Nucleic Acids Res. 2008 Jan;36(Database issue):D102-6. doi: 10.1093/nar/gkm955. Epub 2007 Nov 15. Nucleic Acids Res. 2008. PMID: 18006571 Free PMC article. - JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles.
Castro-Mondragon JA, Riudavets-Puig R, Rauluseviciute I, Lemma RB, Turchi L, Blanc-Mathieu R, Lucas J, Boddie P, Khan A, Manosalva Pérez N, Fornes O, Leung TY, Aguirre A, Hammal F, Schmelter D, Baranasic D, Ballester B, Sandelin A, Lenhard B, Vandepoele K, Wasserman WW, Parcy F, Mathelier A. Castro-Mondragon JA, et al. Nucleic Acids Res. 2022 Jan 7;50(D1):D165-D173. doi: 10.1093/nar/gkab1113. Nucleic Acids Res. 2022. PMID: 34850907 Free PMC article.
Cited by
- A KRAS-responsive long non-coding RNA controls microRNA processing.
Shi L, Magee P, Fassan M, Sahoo S, Leong HS, Lee D, Sellers R, Brullé-Soumaré L, Cairo S, Monteverde T, Volinia S, Smith DD, Di Leva G, Galuppini F, Paliouras AR, Zeng K, O'Keefe R, Garofalo M. Shi L, et al. Nat Commun. 2021 Apr 1;12(1):2038. doi: 10.1038/s41467-021-22337-3. Nat Commun. 2021. PMID: 33795683 Free PMC article. - Protein-DNA binding in high-resolution.
Mahony S, Pugh BF. Mahony S, et al. Crit Rev Biochem Mol Biol. 2015;50(4):269-83. doi: 10.3109/10409238.2015.1051505. Epub 2015 Jun 3. Crit Rev Biochem Mol Biol. 2015. PMID: 26038153 Free PMC article. Review. - High-resolution analysis of differential gene expression during skeletal muscle atrophy and programmed cell death.
Tsuji J, Thomson T, Chan E, Brown CK, Oppenheimer J, Bigelow C, Dong X, Theurkauf WE, Weng Z, Schwartz LM. Tsuji J, et al. Physiol Genomics. 2020 Oct 1;52(10):492-511. doi: 10.1152/physiolgenomics.00047.2020. Epub 2020 Sep 14. Physiol Genomics. 2020. PMID: 32926651 Free PMC article. - PAX6 MiniPromoters drive restricted expression from rAAV in the adult mouse retina.
Hickmott JW, Chen CY, Arenillas DJ, Korecki AJ, Lam SL, Molday LL, Bonaguro RJ, Zhou M, Chou AY, Mathelier A, Boye SL, Hauswirth WW, Molday RS, Wasserman WW, Simpson EM. Hickmott JW, et al. Mol Ther Methods Clin Dev. 2016 Aug 10;3:16051. doi: 10.1038/mtm.2016.51. eCollection 2016. Mol Ther Methods Clin Dev. 2016. PMID: 27556059 Free PMC article. - A Systems View of the Differences between APOE ε4 Carriers and Non-carriers in Alzheimer's Disease.
Jiang S, Tang L, Zhao N, Yang W, Qiu Y, Chen HZ. Jiang S, et al. Front Aging Neurosci. 2016 Jul 12;8:171. doi: 10.3389/fnagi.2016.00171. eCollection 2016. Front Aging Neurosci. 2016. PMID: 27462267 Free PMC article.
References
- Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 2004;5:276–287. - PubMed
- Stormo GD. DNA binding sites: representation and discovery. Bioinformatics. 2000;16:16–23. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources