Adenovirus detection by the cGAS/STING/TBK1 DNA sensing cascade - PubMed (original) (raw)
Adenovirus detection by the cGAS/STING/TBK1 DNA sensing cascade
Eric Lam et al. J Virol. 2014 Jan.
Abstract
Adenovirus (Ad) infection triggers a cell-specific antiviral response following exposure of viral DNA to the intracellular compartment. A variety of DNA sensors (DAI, AIM2, DDx41, RNA polymerase [Pol] III, and IFI16 [p204]) have been identified in recent years; however, the DNA sensor involved in detection of adenovirus has not been established. Cyclic GMP-AMP synthase (cGAS), a DNA sensor that produces a cyclic guanine-adenine dinucleotide (cGAMP) inducer of STING, has been examined to determine its role in generating an antiadenoviral response. Short hairpin RNA (shRNA) lentiviral vectors targeting TBK1, STING, and cGAS were established in murine MS1 endothelial and RAW 264.7 macrophage cell lines. Knockdown of TBK1, STING, and cGAS results in a dramatic reduction in the activation of the primary antiviral response marker phosphorylated interferon (IFN) response factor 3 (IRF3) following exposure to adenovirus. Furthermore, activation of secondary type I IFN signaling targets ((ptyr)STAT1 and (ptyr)STAT2 [(ptyr)STAT1/2]) was also compromised. Consistent with compromised activation of primary and secondary response markers, transcriptional activation of IRF3-responsive genes (beta IFN [IFN-β], ISG15, ISG54) and secondary response transcripts were diminished in cells knocked down in cGAS, STING, or TBK1. These data establish cGAS as the dominant cytosolic DNA sensor responsible for detection of internalized adenovirus leading to induction of the type I interferon antiviral cascade.
Figures
FIG 1
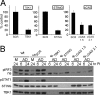
shRNA knockdown of cGAS, TBK1, and STING in MS1 endothelial cells. (A) RT-qPCR quantitation of mRNA levels in MS1 endothelial cells after knockdown and puromycin selection using shRNA lentiviral vectors targeting TBK1, STING, and cGAS. Samples were normalized to the cellular control TBP. One shSCR control sample was selected as representing a 100% standard. All qRT-PCR assays included biological triplicates as well as technical triplicates of each sample. (B) Western analysis of lysates harvested from wild-type (wt) or stable shRNA MS1 knockdown cell line pools at 6 or 24 h post-mock or -Ad5CiG infection using the indicated antibody. ns corresponds to a nonspecific band. M corresponds to mock treatment and AD to Ad5CiG infection. All experiments were carried out a minimum of three times; representative results are presented.
FIG 2
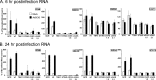
Comparative RT-qPCR of RNA isolated from shSCR, cGAS, and STING and TBK1 shRNA knockdown MS1 endothelial cell pools infected with adenovirus. Data represent the results of two-step RT-qPCR of RNA isolated from mock- or Ad5CiG-infected MS1 and shRNA knockdown cell pools harvested at 6 (A) and 24 (B) h pi. Results are shown for PCR primers corresponding to IFN-β-, ISG15-, ISG54-, and STAT1-inducible transcripts. All samples were normalized to TBP using the ΔΔ_CT_ method as described in Materials and Methods. The value for scramble sample 1 was set as an arbitrary unit of 100.
FIG 3
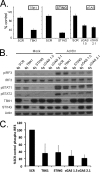
Stable shRNA knockdown of cGAS, TBK1, and STING in the RAW264.7 macrophage cell line. (A) RT-qPCR quantitation of mRNA levels in the RAW264.7 macrophage cell line after knockdown and puromycin selection using shRNA lentiviral vectors targeting TBK1, STING, and cGAS. shSCR corresponds to scramble control shRNA. Samples were normalized to the cellular control TBP. One shSCR control sample was selected as representing a 100% standard. (B) Western analysis of lysates harvested from wild-type or stable shRNA macrophage knockdown cell line pools 6 h post-Ad5CiG infection using the indicated antibody. (C) Quantitation of AdV-induced phospho-IRF3 for each sh knockdown. Determinations were averaged from the results of 5 or more experiments (as described for panel B), where the shSCR value was set as 100% for each experiment.
FIG 4
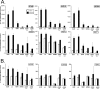
Comparative RT-qPCR of RNA isolated from shSCR, cGAS, STING, and TBK1 shRNA knockdown RAW264.7 shRNA cell pools infected with adenovirus. Data represent the results of two-step RT-qPCR of RNA isolated from mock- or Ad5CiG-infected RAW264.7 cell lines and shRNA knockdown cell pools harvested at 6 h pi. Results are shown for PCR primers corresponding to primary response genes IFN-β, ISG15, and ISG54 and secondary response-inducible transcripts STAT1, TNF-α, and IRF7 (A) and to shRNA-targeted genes (B). All samples were normalized to TBP using the ΔΔ_CT_ method as described in Materials and Methods. The value for scramble sample 1 was set as an arbitrary unit of 100.
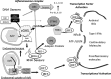
FIG 5
Summary model for rAdV DNA sensing. Following endosomal entry of rAdV into the cell (lower left), viral DNA can trigger the TLR9 pathway, which through the MyD88 adaptor leads to activation of IRF7 and IRF5 but not IRF3. Alternatively, adenovirus escapes the endosome, revealing virus and viral DNA complexes to the cytosolic compartment. An array of DNA sensors are available to bind viral DNA; one DNA sensor, cyclic GMP-AMP synthase (cGAS), has been shown to influence the antiviral response to adenovirus infection in murine endothelial and macrophage cell lines. DNA engagement by cGAS results in production of a unique cGAMP containing 2′5′ and 3′5 linkages that bind to the adaptor STING and leads to TBK1 activation. Active TBK1 mediates C-terminal phosphorylation of IRF3. IRF3 dimerizes, translocates to the nucleus (indicated by the double-line structure), and associates with transcription units (IFN-β or ISG15, -54, and -56). In collaboration with additional transcription factors (NF-κB, ATF/cJUN), gene expression is induced. AIM2 and p204 are Hin-200 proteins that complex with ASC through pyrin domains to form an inflammasome complex, leading to secretion of IL-1 and IL-18. DNA template present either in the cytosol or in the nucleus can be transcribed. Small viral RNAs have been implicated in stimulating the antiviral response through activation of RNA sensors (MDA-5 and RIG-I) which in turn complex with IPS (MAVS) and activate TBK1.
Similar articles
- Unabated adenovirus replication following activation of the cGAS/STING-dependent antiviral response in human cells.
Lam E, Falck-Pedersen E. Lam E, et al. J Virol. 2014 Dec;88(24):14426-39. doi: 10.1128/JVI.02608-14. Epub 2014 Oct 8. J Virol. 2014. PMID: 25297994 Free PMC article. - Sensing adenovirus infection: activation of interferon regulatory factor 3 in RAW 264.7 cells.
Stein SC, Falck-Pedersen E. Stein SC, et al. J Virol. 2012 Apr;86(8):4527-37. doi: 10.1128/JVI.07071-11. Epub 2012 Feb 15. J Virol. 2012. PMID: 22345436 Free PMC article. - Diminished Innate Antiviral Response to Adenovirus Vectors in cGAS/STING-Deficient Mice Minimally Impacts Adaptive Immunity.
Anghelina D, Lam E, Falck-Pedersen E. Anghelina D, et al. J Virol. 2016 Jun 10;90(13):5915-27. doi: 10.1128/JVI.00500-16. Print 2016 Jul 1. J Virol. 2016. PMID: 27076643 Free PMC article. - The mechanism of double-stranded DNA sensing through the cGAS-STING pathway.
Shu C, Li X, Li P. Shu C, et al. Cytokine Growth Factor Rev. 2014 Dec;25(6):641-8. doi: 10.1016/j.cytogfr.2014.06.006. Epub 2014 Jun 22. Cytokine Growth Factor Rev. 2014. PMID: 25007740 Free PMC article. Review. - The molecular mechanism of dsDNA sensing through the cGAS-STING pathway.
Shinde O, Li P. Shinde O, et al. Adv Immunol. 2024;162:1-21. doi: 10.1016/bs.ai.2024.02.003. Epub 2024 Mar 2. Adv Immunol. 2024. PMID: 38866436 Review.
Cited by
- Mechanism and Application Prospects of NLRC3 Regulating cGAS-STING Pathway in Lung Cancer Immunotherapy.
Wang Q, Ren Z, Zhao J, Zheng T, Tong L, Liu J, Dai Z, Tang S. Wang Q, et al. Int J Med Sci. 2024 Oct 7;21(13):2613-2622. doi: 10.7150/ijms.102328. eCollection 2024. Int J Med Sci. 2024. PMID: 39439455 Free PMC article. Review. - Empowering brain tumor management: chimeric antigen receptor macrophage therapy.
Feng F, Shen J, Qi Q, Zhang Y, Ni S. Feng F, et al. Theranostics. 2024 Sep 3;14(14):5725-5742. doi: 10.7150/thno.98290. eCollection 2024. Theranostics. 2024. PMID: 39310093 Free PMC article. Review. - The Immune System-A Double-Edged Sword for Adenovirus-Based Therapies.
Wallace R, Bliss CM, Parker AL. Wallace R, et al. Viruses. 2024 Jun 17;16(6):973. doi: 10.3390/v16060973. Viruses. 2024. PMID: 38932265 Free PMC article. Review. - Biological features of fowl adenovirus serotype-4.
Rashid F, Xie Z, Wei Y, Xie Z, Xie L, Li M, Luo S. Rashid F, et al. Front Cell Infect Microbiol. 2024 Jun 10;14:1370414. doi: 10.3389/fcimb.2024.1370414. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38915924 Free PMC article. Review. - Nervous necrosis virus capsid protein and Protein A dynamically modulate the fish cGAS-mediated IFN signal pathway to facilitate viral evasion.
Huang S, Yang L, Zheng R, Weng S, He J, Xie J. Huang S, et al. J Virol. 2024 Jul 23;98(7):e0068624. doi: 10.1128/jvi.00686-24. Epub 2024 Jun 18. J Virol. 2024. PMID: 38888343 Free PMC article.
References
- Fejer G, Drechsel L, Liese J, Schleicher U, Ruzsics Z, Imelli N, Greber UF, Keck S, Hildenbrand B, Krug A, Bogdan C, Freudenberg MA. 2008. Key role of splenic myeloid DCs in the IFN-alphabeta response to adenoviruses in vivo. PLoS Pathog. 4:e1000208. 10.1371/journal.ppat.1000208 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous