Beyond GWASs: illuminating the dark road from association to function - PubMed (original) (raw)
Review
Beyond GWASs: illuminating the dark road from association to function
Stacey L Edwards et al. Am J Hum Genet. 2013.
Abstract
Genome-wide association studies (GWASs) have enabled the discovery of common genetic variation contributing to normal and pathological traits and clinical drug responses, but recognizing the precise targets of these associations is now the major challenge. Here, we review recent approaches to the functional follow-up of GWAS loci, including fine mapping of GWAS signal(s), prioritization of putative functional SNPs by the integration of genetic epidemiological and bioinformatic methods, and in vitro and in vivo experimental verification of predicted molecular mechanisms for identifying the targeted genes. The majority of GWAS-identified variants fall in noncoding regions of the genome. Therefore, this review focuses on strategies for assessing likely mechanisms affected by noncoding variants; such mechanisms include transcriptional regulation, noncoding RNA function, and epigenetic regulation. These approaches have already accelerated progress from genetic studies to biological knowledge and might ultimately guide the development of prognostic, preventive, and therapeutic measures.
Copyright © 2013 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures
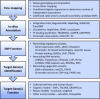
Figure 1
Workflow for Functionally Analyzing and Interpreting GWAS Loci
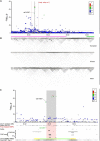
Figure 2
Integrated Genetic and Genomic Data at the 11q13 Breast Cancer Susceptibility Locus (A) Manhattan plot displaying the strength of genetic association (−log10 p) versus chromosomal position (Mb). Each dot represents a genotyped or imputed SNP. Dot colors signify the degree of pairwise correlation (r2) with the top SNP, as presented in the color key. White dots depict SNPs for which r2 values are unknown. The gray shaded stripe represents the iCHAV, encompassing a physical area bound by SNPs that are statistically indistinguishable by stepwise conditional analysis and including the GWAS lead SNP rs614367. The purple dotted line represents the threshold for genome-wide significance (p = 5 × 10−8). (B) Linkage disequilibrium plots depicting pairwise correlation between SNPs genotyped in the 1000 Genomes Project for European (CEU), African (YRI), and Asian (CHB) populations. The plots are in grayscale, for which white and black signify r2 = 0 and 1, respectively. The pink and green bars denote haplotype blocks described in the text in relation to transethnic fine mapping. (C) Inset from panel (A). The UCSC Genome Browser was used for visualizing ENCODE data tracks, which are indicative of regulatory function. The pink stripe indicates the genomic region corresponding to the iCHAV at 11q13, and the locations of the fine-mapped SNPs are shown as red marks. Regions of open chromatin, indicative of putative regulatory signals, are detected as DNaseI hypersensitive sites (DHSs) and are marked. ChIP-seq data for histone marks associated with regulatory regions and specific TFs relevant to breast cancer are shown. The ENCODE ChromHMM track represents integrated analysis of chromatin states based upon histone ChIP-seq data from human mammary epithelial cells (HMECs). Color coding is as follows: green, weak transcription; yellow and orange, enhancer; red, promoter; blue, insulator; gray, repressed. RNA Pol II ChIA-PET data from MCF7 cells are represented as a grayscale bar; darker regions indicate more frequent interactions.
Similar articles
- A practical view of fine-mapping and gene prioritization in the post-genome-wide association era.
Broekema RV, Bakker OB, Jonkers IH. Broekema RV, et al. Open Biol. 2020 Jan;10(1):190221. doi: 10.1098/rsob.190221. Epub 2020 Jan 15. Open Biol. 2020. PMID: 31937202 Free PMC article. - Lessons from postgenome-wide association studies: functional analysis of cancer predisposition loci.
Monteiro AN, Freedman ML. Monteiro AN, et al. J Intern Med. 2013 Nov;274(5):414-24. doi: 10.1111/joim.12085. J Intern Med. 2013. PMID: 24127939 Free PMC article. Review. - Fine-Mapping of Common Genetic Variants Associated with Colorectal Tumor Risk Identified Potential Functional Variants.
Du M, Jiao S, Bien SA, Gala M, Abecasis G, Bezieau S, Brenner H, Butterbach K, Caan BJ, Carlson CS, Casey G, Chang-Claude J, Conti DV, Curtis KR, Duggan D, Gallinger S, Haile RW, Harrison TA, Hayes RB, Hoffmeister M, Hopper JL, Hudson TJ, Jenkins MA, Küry S, Le Marchand L, Leal SM, Newcomb PA, Nickerson DA, Potter JD, Schoen RE, Schumacher FR, Seminara D, Slattery ML, Hsu L, Chan AT, White E, Berndt SI, Peters U. Du M, et al. PLoS One. 2016 Jul 5;11(7):e0157521. doi: 10.1371/journal.pone.0157521. eCollection 2016. PLoS One. 2016. PMID: 27379672 Free PMC article. - Disease-Associated Single-Nucleotide Polymorphisms From Noncoding Regions in Juvenile Idiopathic Arthritis Are Located Within or Adjacent to Functional Genomic Elements of Human Neutrophils and CD4+ T Cells.
Jiang K, Zhu L, Buck MJ, Chen Y, Carrier B, Liu T, Jarvis JN. Jiang K, et al. Arthritis Rheumatol. 2015 Jul;67(7):1966-77. doi: 10.1002/art.39135. Arthritis Rheumatol. 2015. PMID: 25833190 Free PMC article. - Molecular mechanisms underlying noncoding risk variations in psychiatric genetic studies.
Xiao X, Chang H, Li M. Xiao X, et al. Mol Psychiatry. 2017 Apr;22(4):497-511. doi: 10.1038/mp.2016.241. Epub 2017 Jan 3. Mol Psychiatry. 2017. PMID: 28044063 Free PMC article. Review.
Cited by
- Research Priorities for Endometriosis.
Rogers PA, Adamson GD, Al-Jefout M, Becker CM, D'Hooghe TM, Dunselman GA, Fazleabas A, Giudice LC, Horne AW, Hull ML, Hummelshoj L, Missmer SA, Montgomery GW, Stratton P, Taylor RN, Rombauts L, Saunders PT, Vincent K, Zondervan KT; WES/WERF Consortium for Research Priorities in Endometriosis. Rogers PA, et al. Reprod Sci. 2017 Feb;24(2):202-226. doi: 10.1177/1933719116654991. Epub 2016 Sep 27. Reprod Sci. 2017. PMID: 27368878 Free PMC article. - Genetic Variation Determines PPARγ Function and Anti-diabetic Drug Response In Vivo.
Soccio RE, Chen ER, Rajapurkar SR, Safabakhsh P, Marinis JM, Dispirito JR, Emmett MJ, Briggs ER, Fang B, Everett LJ, Lim HW, Won KJ, Steger DJ, Wu Y, Civelek M, Voight BF, Lazar MA. Soccio RE, et al. Cell. 2015 Jul 2;162(1):33-44. doi: 10.1016/j.cell.2015.06.025. Cell. 2015. PMID: 26140591 Free PMC article. - A Human-Specific Schizophrenia Risk Tandem Repeat Affects Alternative Splicing of a Human-Unique Isoform AS3MTd2d3 and Mushroom Dendritic Spine Density.
Cai X, Yang ZH, Li HJ, Xiao X, Li M, Chang H. Cai X, et al. Schizophr Bull. 2021 Jan 23;47(1):219-227. doi: 10.1093/schbul/sbaa098. Schizophr Bull. 2021. PMID: 32662510 Free PMC article. - Transcriptome-wide association study and eQTL colocalization identify potentially causal genes responsible for human bone mineral density GWAS associations.
Al-Barghouthi BM, Rosenow WT, Du KP, Heo J, Maynard R, Mesner L, Calabrese G, Nakasone A, Senwar B, Gerstenfeld L, Larner J, Ferguson V, Ackert-Bicknell C, Morgan E, Brautigan D, Farber CR. Al-Barghouthi BM, et al. Elife. 2022 Nov 23;11:e77285. doi: 10.7554/eLife.77285. Elife. 2022. PMID: 36416764 Free PMC article. - Most brain disease-associated and eQTL haplotypes are not located within transcription factor DNase-seq footprints in brain.
Handel AE, Gallone G, Zameel Cader M, Ponting CP. Handel AE, et al. Hum Mol Genet. 2017 Jan 1;26(1):79-89. doi: 10.1093/hmg/ddw369. Hum Mol Genet. 2017. PMID: 27798116 Free PMC article.
References
- Hindorff, L.A., MacArthur, J., Morales, J., Junkins, H.A., Hall, P.N., Klemm, A.K., and Manolio, T.A. A Catalog of Published Genome-Wide Association Studies. http://www.genome.gov/gwastudies.
- Kruglyak L. Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat. Genet. 1999;22:139–144. - PubMed
- Cooper G.M., Shendure J. Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nat. Rev. Genet. 2011;12:628–640. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 16565/CRUK_/Cancer Research UK/United Kingdom
- C8197/A10123/CRUK_/Cancer Research UK/United Kingdom
- C8197/A10865/CRUK_/Cancer Research UK/United Kingdom
- C1287/A10118/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources