Structural interactions between inhibitor and substrate docking sites give insight into mechanisms of human PS1 complexes - PubMed (original) (raw)
. 2014 Jan 7;22(1):125-35.
doi: 10.1016/j.str.2013.09.018. Epub 2013 Nov 7.
Stephen Hsueh-Jeng Lu 1, Ching-Ju Tsai 1, Christopher Bohm 2, Seema Qamar 1, Roger B Dodd 1, William Meadows 1, Amy Jeon 2, Adam McLeod 2, Fusheng Chen 2, Muriel Arimon 3, Oksana Berezovska 3, Bradley T Hyman 3, Taisuke Tomita 4, Takeshi Iwatsubo 4, Christopher M Johnson 5, Lindsay A Farrer 6, Gerold Schmitt-Ulms 2, Paul E Fraser 2, Peter H St George-Hyslop 7
Affiliations
- PMID: 24210759
- PMCID: PMC3887256
- DOI: 10.1016/j.str.2013.09.018
Structural interactions between inhibitor and substrate docking sites give insight into mechanisms of human PS1 complexes
Yi Li et al. Structure. 2014.
Abstract
Presenilin-mediated endoproteolysis of transmembrane proteins plays a key role in physiological signaling and in the pathogenesis of Alzheimer disease and some cancers. Numerous inhibitors have been found via library screens, but their structural mechanisms remain unknown. We used several biophysical techniques to investigate the structure of human presenilin complexes and the effects of peptidomimetic γ-secretase inhibitors. The complexes are bilobed. The head contains nicastrin ectodomain. The membrane-embedded base has a central channel and a lateral cleft, which may represent the initial substrate docking site. Inhibitor binding induces widespread structural changes, including rotation of the head and closure of the lateral cleft. These changes block substrate access to the catalytic pocket and inhibit the enzyme. Intriguingly, peptide substrate docking has reciprocal effects on the inhibitor binding site. Similar reciprocal shifts may underlie the mechanisms of other inhibitors and of the "lateral gate" through which substrates access to the catalytic site.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
Graphical abstract
Figure 1
Compound E Binding Induces Conformational Changes in PS1 Complexes that Prevent Detergent-Mediated Dissociation of PS1 Complexes into Hemi-Complexes (A) Cartoon depicting the hemi-complexes. Substrates have been previously shown to bind to both PS1-NTF and PS1-CTF. (B) In 0.1% DDM, nicastrin coimmunoprecipitates all complex components: PS1-NTF, PS1-CTF, aph1, and pen2. With increasing detergent concentration, nicastrin coimmunoprecipitates only aph1 and PS1-CTF. (C) Incubation of complexes with compound E stabilizes complexes across a range of detergent concentrations. (D) The compound E-mediated stabilization of the PS1 complexes is dose dependent.
Figure 2
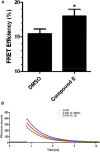
FLIM-FRET Analysis of Purified, Mature, Catalytically Active, GFP- and RFP-Tagged PS1 Complexes Confirms that Compound E Binding Causes Conformational Changes (A) FRET efficiencies of GFP/RFP-tagged PS1 complexes are improved after incubation in 10 μM compound E (DMSO control: 15.46 ± 0.69% mean ± SEM; 10 μM compound E: 18.02 ± 0.95%, n = three independent experiments). ∗p ≤ 0.05. (B) Representative fluorescence decay curves for donor GFP fluorescence under each experimental condition. G-PS1 and G-PS1-R decay curves and fluorescence lifetimes were derived from fitting the experimentally observed photon counts by single exponential decay and double exponential decay models, respectively. The relationship of the FRET donor-receiver pair on the two hemi-complexes is displayed in Figure S1A. A western blot demonstrating complete endoproteolysis and maturation of the GFP/RFP-tagged PS1 complexes is displayed in Figure S1B.
Figure 3
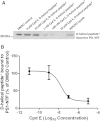
Noncleavable Peptidic D-Helical Substrate Mimic and Compound E Have Reciprocal Allosteric Effects on Each Other’s Binding to PS1 Complexes and Compound E Inhibits Binding of the D-Amino-Acid Helical Substrate to the Initial Substrate Docking Site of PS1 Complexes (A) A representative blot showing progressive inhibition of D-helical photoprobe binding to PS1-NTF in the presence of increasing concentrations of compound E. (B) Quantitative results of four independent experiments expressed as percentage of DMSO control. Error bars are SEM.
Figure 4
Noncleavable Peptidic D-Helical Substrate Mimic and Compound E Have Reciprocal Allosteric Effects on Each Other’s Binding to PS1 Complexes Preincubation of the PS1 complexes with the D-helical substrate mimic enhanced binding of compound E to the PS1 complex.
Figure 5
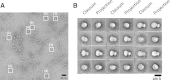
Raw Particles and 3D Model Validation of Human PS1 Complex (A) CCD image of native PS1 complexes. Representative particle shapes are highlighted by white boxes; boxes 1–3, bilobed shapes; boxes 4–8, round or oval shapes; boxes 6–8 show suggestive central cavities. Scale bar, 20 nm. (B) Classums of native PS1 particles compared with 2D projections of the final model for native PS1 complexes. The classums and corresponding 2D projections are highly similar in size, shape, and internal density distribution. Bilobed, oval, and round shapes are seen that are similar to the raw particle images. Central cavities in the base domain are apparent in most classum/2D-projection pairs. Scale bar, 100 Å. Supplemental information is available, including a detailed flowchart of the complex purification algorithm (Figure S2), silver-stained SDS-PAGE gel showing the presence of all complex components and blue native PAGE showing their monodispersity and catalytic activity that can be inhibited by compound E (Figure S3), mass analysis of the complex using size exclusion chromatography with SEC-MALS (Figure S4), detailed flowchart of the model-building algorithm (Figure S5), and additional classum images (Figure S6).
Figure 6
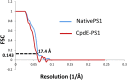
Resolution of the Final Maps as Evaluated by gsFSC Method The 0.143 threshold resolution of both the native and compound E-bound PS1 map was 17.4 Å resolution.
Figure 7
Top-Down Vertical, Lateral, and Cross-Sectional Views of 3D Reconstructions of PS1 Complexes Reveal that Both Native PS1 Complexes and Compound E-Bound Complexes Have an Irregular Bilobed Shape (A) The native PS1 complex contains a head domain and a base domain. The base domain has a lateral cleft and central cavity/channel, which appears to open onto the upper/extracellular surface and also onto the lower surface via a smaller pore. The shaded lipid bilayer represents the boundaries of a putative membrane. (B) 3D reconstructions of compound E-bound PS1 complexes reveal a similar structure, with the rotation and tilting of the head. Density shifts on the external surface of the base result in closure of the lateral cleft and of the lower pore of central channel. (C) Corresponding vertical and lateral views of the difference map, which was calculated using the UCSF Chimera package. Blue mesh is the native PS1 complex, and the pink mesh is compound E-bound PS1. Positive density is represented in green. Negative density is displayed in red. A detailed flowchart of the model-building algorithm is available in Figure S5. A rotating animated video of the native complex built in chimera (Movie S1) and an animated video comparing the native and compound E complexes (Movie S2) are available in online supplemental data files.
Figure 8
Antibodies to the Ectodomain of Nicastrin Label the Head Domain, Indicating that the Head Domain Is Lumenal/Extracellular Representative class average images of native and anti-nicastrin antibody-labeled PS1 complexes reveal that anti-nicastrin antibody-labeled complexes have an increased density of the head domain. The box width is 261.12 Å.
Similar articles
- Structural biology of presenilin 1 complexes.
Li Y, Bohm C, Dodd R, Chen F, Qamar S, Schmitt-Ulms G, Fraser PE, St George-Hyslop PH. Li Y, et al. Mol Neurodegener. 2014 Dec 18;9:59. doi: 10.1186/1750-1326-9-59. Mol Neurodegener. 2014. PMID: 25523933 Free PMC article. Review. - Mutation analysis of the presenilin 1 N-terminal domain reveals a broad spectrum of gamma-secretase activity toward amyloid precursor protein and other substrates.
Gong P, Vetrivel KS, Nguyen PD, Meckler X, Cheng H, Kounnas MZ, Wagner SL, Parent AT, Thinakaran G. Gong P, et al. J Biol Chem. 2010 Dec 3;285(49):38042-52. doi: 10.1074/jbc.M110.132613. Epub 2010 Oct 4. J Biol Chem. 2010. PMID: 20921220 Free PMC article. - Allosteric Modulation of Intact γ-Secretase Structural Dynamics.
Lee JY, Feng Z, Xie XQ, Bahar I. Lee JY, et al. Biophys J. 2017 Dec 19;113(12):2634-2649. doi: 10.1016/j.bpj.2017.10.012. Biophys J. 2017. PMID: 29262358 Free PMC article. - Identification of presenilin 1-selective γ-secretase inhibitors with reconstituted γ-secretase complexes.
Lee J, Song L, Terracina G, Bara T, Josien H, Asberom T, Sasikumar TK, Burnett DA, Clader J, Parker EM, Zhang L. Lee J, et al. Biochemistry. 2011 Jun 7;50(22):4973-80. doi: 10.1021/bi200026m. Epub 2011 May 13. Biochemistry. 2011. PMID: 21528914 - The Alzheimer's disease-associated gamma-secretase complex: functional domains in the presenilin 1 protein.
Laudon H, Winblad B, Näslund J. Laudon H, et al. Physiol Behav. 2007 Sep 10;92(1-2):115-20. doi: 10.1016/j.physbeh.2007.05.037. Epub 2007 May 21. Physiol Behav. 2007. PMID: 17588625 Review.
Cited by
- Suppressor Mutations for Presenilin 1 Familial Alzheimer Disease Mutants Modulate γ-Secretase Activities.
Futai E, Osawa S, Cai T, Fujisawa T, Ishiura S, Tomita T. Futai E, et al. J Biol Chem. 2016 Jan 1;291(1):435-46. doi: 10.1074/jbc.M114.629287. Epub 2015 Nov 11. J Biol Chem. 2016. PMID: 26559975 Free PMC article. - Current and future implications of basic and translational research on amyloid-β peptide production and removal pathways.
Bohm C, Chen F, Sevalle J, Qamar S, Dodd R, Li Y, Schmitt-Ulms G, Fraser PE, St George-Hyslop PH. Bohm C, et al. Mol Cell Neurosci. 2015 May;66(Pt A):3-11. doi: 10.1016/j.mcn.2015.02.016. Epub 2015 Mar 4. Mol Cell Neurosci. 2015. PMID: 25748120 Free PMC article. Review. - Role of cholesterol in substrate recognition by [Formula: see text]-secretase.
Nierzwicki Ł, Olewniczak M, Chodnicki P, Czub J. Nierzwicki Ł, et al. Sci Rep. 2021 Jul 26;11(1):15213. doi: 10.1038/s41598-021-94618-2. Sci Rep. 2021. PMID: 34312439 Free PMC article. - The Canonical Notch Signaling Pathway: Structural and Biochemical Insights into Shape, Sugar, and Force.
Kovall RA, Gebelein B, Sprinzak D, Kopan R. Kovall RA, et al. Dev Cell. 2017 May 8;41(3):228-241. doi: 10.1016/j.devcel.2017.04.001. Dev Cell. 2017. PMID: 28486129 Free PMC article. Review. - Alzheimer's disease-associated mutations increase amyloid precursor protein resistance to γ-secretase cleavage and the Aβ42/Aβ40 ratio.
Xu TH, Yan Y, Kang Y, Jiang Y, Melcher K, Xu HE. Xu TH, et al. Cell Discov. 2016 Aug 23;2:16026. doi: 10.1038/celldisc.2016.26. eCollection 2016. Cell Discov. 2016. PMID: 27625790 Free PMC article.
References
- Bürckstümmer T., Bennett K.L., Preradovic A., Schütze G., Hantschel O., Superti-Furga G., Bauch A. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat. Methods. 2006;3:1013–1019. - PubMed
- Burgard, C. (2009). Structural and functional characterization of components of the ER protein translocase. PhD thesis, University of The Saarland, Saarbrücken, Germany.
- Chen F., Hasegawa H., Schmitt-Ulms G., Kawarai T., Bohm C., Katayama T., Gu Y., Sanjo N., Glista M., Rogaeva E. TMP21 is a presenilin complex component that modulates gamma-secretase but not epsilon-secretase activity. Nature. 2006;440:1208–1212. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CAPMC/ CIHR/Canada
- P30 AG013846/AG/NIA NIH HHS/United States
- 089703/WT_/Wellcome Trust/United Kingdom
- MC_G1000734/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- 100140/WT_/Wellcome Trust/United Kingdom
- 081864/WT_/Wellcome Trust/United Kingdom
- P01 AG015379/AG/NIA NIH HHS/United States
- AG15379/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources