Molecular and Epigenetic Mechanisms of MLL in Human Leukemogenesis - PubMed (original) (raw)
Molecular and Epigenetic Mechanisms of MLL in Human Leukemogenesis
Erica Ballabio et al. Cancers (Basel). 2012.
Abstract
Epigenetics is often defined as the study of heritable changes in gene expression or chromosome stability that don't alter the underlying DNA sequence. Epigenetic changes are established through multiple mechanisms that include DNA methylation, non-coding RNAs and the covalent modification of specific residues on histone proteins. It is becoming clear not only that aberrant epigenetic changes are common in many human diseases such as leukemia, but that these changes by their very nature are malleable, and thus are amenable to treatment. Epigenetic based therapies have so far focused on the use of histone deacetylase (HDAC) inhibitors and DNA methyltransferase inhibitors, which tend to have more general and widespread effects on gene regulation in the cell. However, if a unique molecular pathway can be identified, diseases caused by epigenetic mechanisms are excellent candidates for the development of more targeted therapies that focus on specific gene targets, individual binding domains, or specific enzymatic activities. Designing effective targeted therapies depends on a clear understanding of the role of epigenetic mutations during disease progression. The Mixed Lineage Leukemia (MLL) protein is an example of a developmentally important protein that controls the epigenetic activation of gene targets in part by methylating histone 3 on lysine 4. MLL is required for normal development, but is also mutated in a subset of aggressive human leukemias and thus provides a useful model for studying the link between epigenetic cell memory and human disease. The most common MLL mutations are chromosome translocations that fuse the MLL gene in frame with partner genes creating novel fusion proteins. In this review, we summarize recent work that argues MLL fusion proteins could function through a single molecular pathway, but we also highlight important data that suggests instead that multiple independent mechanisms underlie MLL mediated leukemogenesis.
Figures
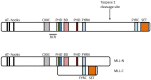
Figure 1
MLL protein structure. MLL has 3 HMG-like AT hooks (black bars) that bind AT rich DNA, a CXXC domain (grey bar) that binds unmethylated CpG DNA, four PHD (Plant Homeo Domain) fingers (yellow, green blue and red bars) that mediate interactions with several proteins; an atypical bromodomain (purple bar), FYRN and FYRC domains (light blue bars) and a C terminal SET domain (orange bar) that methylates histone H3 on lysine 4. Wild type MLL is cleaved by taspase 1 into two fragments: MLL-N and MLL-C. These fragments dimerize to form a stable complex. BCR = breakpoint common region. Adapted from [69].
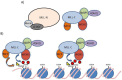
Figure 2
MLL multiprotein complex. (A) MLL is present in the cell as part of a large protein complex. MLL-C is associated with the WDR5, RBBP5, ASH2L and MOF proteins. MLL-N interacts with a wide range of different proteins, for simplicity only menin and LEDGF are shown; (B) The model proposed by Wysocka et al. and Dou et al.: WDR5 recruits the MLL complex to H3K4me2 allowing the addition of another methyl group to histone H3 lysine 4 and acetyl group to histone H4 lysine 16. Additional MLL complexes are then recruited to target genes or existing MLL complexes are transferred to the next nucleosome [95,114]. The yellow triangle represents an acetyl mark, while red circles represent methyl marks. Figure adapted from [114].
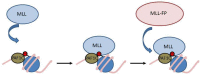
Figure 3
Steps in recruiting MLL and MLL-FPs to the HOXA9 locus. MLL is recruited to a target gene through interactions with the PAF1 complex and H3K4Me3 which results in increased activation of the locus and a more open chromatin conformation. This allows MLL-FP to bind by interactions with PAF1C and CpG rich DNA. Red circles represent methyl marks.
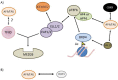
Figure 4
Interactions amongst Super Elongation Complex components (A) AF4, AFF4, AF9, ENL, pTEFb, DOT1L, ELL and EAF protein complexes are linked together by a series of different interactions. BRD4 and PAF1C are also linked to the SEC via interactions with pTEFb and AF9/ENL, respectively. Recent data have also suggested the possibility that AF9/ENL might bind TFIID; (B) AF9/ENL interacts with the H3K79 methyltransferase DOT1L in a complex that excludes other interacting partners.
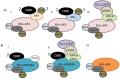
Figure 5
MLL-FPs are present in the cell as part of distinct multiprotein complexes. (A) The MLL-AF4 fusion protein can either bind to AF9 (i) or ENL (ii) or AF4/AFF4 (iii). CBX8 and PAF1C may also be able to interact with wild type AF9 and ENL in the context of the MLL-AF4 complexes, or they may be mutually exclusive (the uncertainty is indicated by a “?”); (B) MLL-AF9 or MLL-ENL can exist in a complex either with DOT1L or with AF4/AFF4/pTEFb/ELL/EAF. CBX8 may be part of these complexes or may be in a distinct, mutually exclusive complex with MLL-ENL or MLL-AF9; (C) MLL-AF6 is the only MLL-FP which doesn’t interact with any SEC component.
Similar articles
- Epigenetic control of gene expression in leukemogenesis: Cooperation between wild type MLL and MLL fusion proteins.
Ballabio E, Milne TA. Ballabio E, et al. Mol Cell Oncol. 2014 Oct 29;1(2):e955330. doi: 10.1080/23723548.2014.955330. eCollection 2014 Apr-Jun. Mol Cell Oncol. 2014. PMID: 27308325 Free PMC article. Review. - Disordered epigenetic regulation in MLL-related leukemia.
Zhang Y, Chen A, Yan XM, Huang G. Zhang Y, et al. Int J Hematol. 2012 Oct;96(4):428-37. doi: 10.1007/s12185-012-1180-0. Epub 2012 Sep 29. Int J Hematol. 2012. PMID: 23054645 Review. - Targeting epigenetic programs in MLL-rearranged leukemias.
Bernt KM, Armstrong SA. Bernt KM, et al. Hematology Am Soc Hematol Educ Program. 2011;2011:354-60. doi: 10.1182/asheducation-2011.1.354. Hematology Am Soc Hematol Educ Program. 2011. PMID: 22160057 Review. - Mouse models of MLL leukemia: recapitulating the human disease.
Milne TA. Milne TA. Blood. 2017 Apr 20;129(16):2217-2223. doi: 10.1182/blood-2016-10-691428. Epub 2017 Feb 8. Blood. 2017. PMID: 28179274 Free PMC article. Review. - The stem cell factor SALL4 is an essential transcriptional regulator in mixed lineage leukemia-rearranged leukemogenesis.
Yang L, Liu L, Gao H, Pinnamaneni JP, Sanagasetti D, Singh VP, Wang K, Mathison M, Zhang Q, Chen F, Mo Q, Rosengart T, Yang J. Yang L, et al. J Hematol Oncol. 2017 Oct 3;10(1):159. doi: 10.1186/s13045-017-0531-y. J Hematol Oncol. 2017. PMID: 28974232 Free PMC article.
Cited by
- Molecular characterization of an MLL1 fusion and its role in chromosomal instability.
Parameswaran S, Vizeacoumar FS, Kalyanasundaram Bhanumathy K, Qin F, Islam MF, Toosi BM, Cunningham CE, Mousseau DD, Uppalapati MC, Stirling PC, Wu Y, Bonham K, Freywald A, Li H, Vizeacoumar FJ. Parameswaran S, et al. Mol Oncol. 2019 Feb;13(2):422-440. doi: 10.1002/1878-0261.12423. Epub 2018 Dec 31. Mol Oncol. 2019. PMID: 30548174 Free PMC article. - [Research Progress of Role and Mechanism of SETD7 in Tumor Occurrence and Progression].
Cao L, Wang M, Xu K. Cao L, et al. Zhongguo Fei Ai Za Zhi. 2023 Jan 20;26(1):38-45. doi: 10.3779/j.issn.1009-3419.2023.106.02. Zhongguo Fei Ai Za Zhi. 2023. PMID: 36792079 Free PMC article. Chinese. - Why are so many MLL lysine methyltransferases required for normal mammalian development?
Crump NT, Milne TA. Crump NT, et al. Cell Mol Life Sci. 2019 Aug;76(15):2885-2898. doi: 10.1007/s00018-019-03143-z. Epub 2019 May 16. Cell Mol Life Sci. 2019. PMID: 31098676 Free PMC article. Review. - H3K79me2/3 controls enhancer-promoter interactions and activation of the pan-cancer stem cell marker PROM1/CD133 in MLL-AF4 leukemia cells.
Godfrey L, Crump NT, O'Byrne S, Lau IJ, Rice S, Harman JR, Jackson T, Elliott N, Buck G, Connor C, Thorne R, Knapp DJHF, Heidenreich O, Vyas P, Menendez P, Inglott S, Ancliff P, Geng H, Roberts I, Roy A, Milne TA. Godfrey L, et al. Leukemia. 2021 Jan;35(1):90-106. doi: 10.1038/s41375-020-0808-y. Epub 2020 Apr 2. Leukemia. 2021. PMID: 32242051 Free PMC article. - Epigenetic control of gene expression in leukemogenesis: Cooperation between wild type MLL and MLL fusion proteins.
Ballabio E, Milne TA. Ballabio E, et al. Mol Cell Oncol. 2014 Oct 29;1(2):e955330. doi: 10.1080/23723548.2014.955330. eCollection 2014 Apr-Jun. Mol Cell Oncol. 2014. PMID: 27308325 Free PMC article. Review.
References
- Akalin A., Garrett-Bakelman F.E., Kormaksson M., Busuttil J., Zhang L., Khrebtukova I., Milne T.A., Huang Y., Biswas D., Hess J.L., et al. Base-pair resolution DNA methylation sequencing reveals profoundly divergent epigenetic landscapes in acute myeloid leukemia. PLoS Genet. 2012;8:e1002781. - PMC - PubMed
LinkOut - more resources
Full Text Sources