International Union of Basic and Clinical Pharmacology. [corrected]. LXXXIX. Update on the extended family of chemokine receptors and introducing a new nomenclature for atypical chemokine receptors - PubMed (original) (raw)
Review
. 2013 Nov 11;66(1):1-79.
doi: 10.1124/pr.113.007724. Print 2014.
Adit Ben-Baruch, Amanda M Burkhardt, Christophe Combadiere, Joshua M Farber, Gerard J Graham, Richard Horuk, Alexander Hovard Sparre-Ulrich, Massimo Locati, Andrew D Luster, Alberto Mantovani, Kouji Matsushima, Philip M Murphy, Robert Nibbs, Hisayuki Nomiyama, Christine A Power, Amanda E I Proudfoot, Mette M Rosenkilde, Antal Rot, Silvano Sozzani, Marcus Thelen, Osamu Yoshie, Albert Zlotnik
Affiliations
- PMID: 24218476
- PMCID: PMC3880466
- DOI: 10.1124/pr.113.007724
Review
International Union of Basic and Clinical Pharmacology. [corrected]. LXXXIX. Update on the extended family of chemokine receptors and introducing a new nomenclature for atypical chemokine receptors
Francoise Bachelerie et al. Pharmacol Rev. 2013.
Erratum in
- Pharmacol Rev. 2014 Apr;66(2):467
Abstract
Sixteen years ago, the Nomenclature Committee of the International Union of Pharmacology approved a system for naming human seven-transmembrane (7TM) G protein-coupled chemokine receptors, the large family of leukocyte chemoattractant receptors that regulates immune system development and function, in large part by mediating leukocyte trafficking. This was announced in Pharmacological Reviews in a major overview of the first decade of research in this field [Murphy PM, Baggiolini M, Charo IF, Hébert CA, Horuk R, Matsushima K, Miller LH, Oppenheim JJ, and Power CA (2000) Pharmacol Rev 52:145-176]. Since then, several new receptors have been discovered, and major advances have been made for the others in many areas, including structural biology, signal transduction mechanisms, biology, and pharmacology. New and diverse roles have been identified in infection, immunity, inflammation, development, cancer, and other areas. The first two drugs acting at chemokine receptors have been approved by the U.S. Food and Drug Administration (FDA), maraviroc targeting CCR5 in human immunodeficiency virus (HIV)/AIDS, and plerixafor targeting CXCR4 for stem cell mobilization for transplantation in cancer, and other candidates are now undergoing pivotal clinical trials for diverse disease indications. In addition, a subfamily of atypical chemokine receptors has emerged that may signal through arrestins instead of G proteins to act as chemokine scavengers, and many microbial and invertebrate G protein-coupled chemokine receptors and soluble chemokine-binding proteins have been described. Here, we review this extended family of chemokine receptors and chemokine-binding proteins at the basic, translational, and clinical levels, including an update on drug development. We also introduce a new nomenclature for atypical chemokine receptors with the stem ACKR (atypical chemokine receptor) approved by the Nomenclature Committee of the International Union of Pharmacology and the Human Genome Nomenclature Committee.
Figures
Fig. 1.
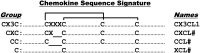
Chemokine primary structure. Chemokines are defined by structure, not function. They are >20% identical for any pairwise protein sequence comparison, and after processing most are 70–80 amino acids long. Four subdivisions are named according to the number and spacing of conserved N-terminal cysteines, as shown; all but three of the human chemokines are in the CXC and CC groups. The cysteines form disulfide bonds as shown by the brackets. Amino acid sequence identity is <30% between members of the four major chemokine groups, but ranges from ∼30 to 99% among members of the same group, indicating separate evolutionary histories. Most chemokine receptors are restricted by group. Most neutrophil-targeted chemokines are in the CXC group, and most monocyte/macrophage-targeted chemokines are in the CC group. Major T and B-cell-targeted chemokines can be found in both groups. The leukocyte target specificity of a chemokine may be narrow or broad and is defined by the expression pattern of its cognate receptor(s).
Fig. 2.
Tertiary structure of chemokines, chemokine receptors, and soluble chemokine-binding proteins. Chemokines have a common fold, and are presented as GAG-tethered molecules on the plasma membrane to leukocytes (upper left [Handel et al., 2005]). The chemokine core, which contains three β sheets arranged in the shape of a Greek key, is overlaid by a C-terminal _α_-helical domain and is flanked by an N-terminal domain that lacks order. Forced chemokine monomers are active but dimer and tetramer structures may occur, and complex quaternary structures bound to GAGs on the surface of cells may be important for function in vivo. Chemokine heterodimers have been described, both CC/CC and CC/CXC. Although in separate groups as defined by cysteine motifs, CXCL16 and CX3CL1 also form a unique multimodular subgroup (upper right [Imai et al., 1997b]). The model shown for these two chemokines depicts a typical chemokine domain, a mucin-like stalk, a transmembrane domain, and a C-terminal cytoplasmic module. They can exist as membrane-bound or cleaved forms, mediating direct G protein-independent cell-cell adhesion and chemotaxis, respectively. Two G protein-coupled chemokine receptors, CXCR1 and CXCR4, have been structurally defined. CXCR4 (lower left) resolves as a dimer (Wu et al., 2010). Atypical chemokine receptors, which do not appear to signal through G proteins, have not yet been defined structurally but are predicted to be 7TM proteins (lower right). Soluble chemokine-binding proteins are produced by microbes (middle left [Alexander et al., 2002]) and invertebrates (middle right [Dias et al., 2009]).
Fig. 3.
The human chemokinome. Genes for chemokines and chemokine receptors each have common ancestors but are distributed on many chromosomes. The two main chemokine gene clusters on chromosomes 4 and 17 (★) contain most of the chemokines that mediate inflammatory responses. Most inflammatory chemokine receptor genes are on chromosomes 2 and 3. Homeostatic chemokine and chemokine receptor genes are scattered on other chromosomes.
Fig. 4.
The human and mouse chemokine gene repertoires are distinct. The syntenic positions of chemokine genes located in clusters are shown schematically and aligned for mouse and human. Chromosome assignments of unclustered genes are listed in the upper box inset. See lower box inset for functional codes. Updated and modified from Nomiyama et al. (2010).
Fig. 5.
Structural relationship of chemokine ligand and receptor proteins in mouse and human. See color code in box inset at the bottom middle. Structures and disease indications are listed above and below, respectively, the names of marketed drugs acting at the three indicated receptors. Dendrograms prepared by S. Tsang, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Fig. 6.
Chemokine receptor specificity for ligands and leukocytes. Abbreviations: Ba, basophil; Ca, cancer; CD4RM, resident memory CD4 T cell; EC, endothelial cell; Eo, eosinophil; Fb, fibroblasts; iDC, immature DC; MC, mast cell; Me, melanocyte; MG, microglial cell; Mo, monocyte; MΦ, macrophage; N, neutrophil; NHC, nonhematopoietic cells; PC, plasma cell; pDC, plasmacytoid DC; Tcm, central memory T cell; Th1, type 1 helper T cell; Tn, naive T cell; eff/mem, effector/memory; thym, thymocytes.
Fig. 7.
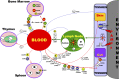
Chemokine receptors important in leukocyte trafficking pathways. Arrows demarcate major leukocyte traffic routes between major tissue and hematopoietic compartments. Cells along the arrows identify some of the cells that follow these routes. The receptors listed for each cell either mark the cell or are used by the cell for trafficking on the route shown. Abbreviations: HSC, hematopoietic stem cell; Tem, effector memory T cell; Teff, effector T cell; Tdp, double positive thymocytes; Tsp, single positive thymocytes; TFH, follicular help T cells.
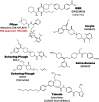
Fig. 8.
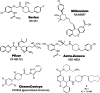
Clinical candidates active at CXCR1 and CXCR2.
Fig. 9.
Clinical candidate active at CXCR3.
Fig. 10.
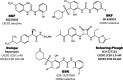
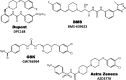
Clinical candidates active at CCR1.
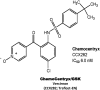
Fig. 11.
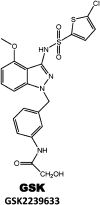
Clinical candidates active at CCR2.
Fig. 12.
Clinical candidates active at CCR3.
Fig. 13.
Clinical candidate active at CCR4.
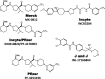
Fig. 14.
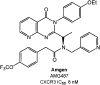
Clinical candidates active at CCR5.
Fig. 15.
Clinical candidate active at CCR9.
Similar articles
- International union of pharmacology. XXII. Nomenclature for chemokine receptors.
Murphy PM, Baggiolini M, Charo IF, Hébert CA, Horuk R, Matsushima K, Miller LH, Oppenheim JJ, Power CA. Murphy PM, et al. Pharmacol Rev. 2000 Mar;52(1):145-76. Pharmacol Rev. 2000. PMID: 10699158 Review. - An atypical addition to the chemokine receptor nomenclature: IUPHAR Review 15.
Bachelerie F, Graham GJ, Locati M, Mantovani A, Murphy PM, Nibbs R, Rot A, Sozzani S, Thelen M. Bachelerie F, et al. Br J Pharmacol. 2015 Aug;172(16):3945-9. doi: 10.1111/bph.13182. Epub 2015 Jun 26. Br J Pharmacol. 2015. PMID: 25958743 Free PMC article. Review. - International Union of Pharmacology. XXX. Update on chemokine receptor nomenclature.
Murphy PM. Murphy PM. Pharmacol Rev. 2002 Jun;54(2):227-9. doi: 10.1124/pr.54.2.227. Pharmacol Rev. 2002. PMID: 12037138 Review. - Overview and potential unifying themes of the atypical chemokine receptor family.
Vacchini A, Locati M, Borroni EM. Vacchini A, et al. J Leukoc Biol. 2016 Jun;99(6):883-92. doi: 10.1189/jlb.2MR1015-477R. Epub 2016 Jan 6. J Leukoc Biol. 2016. PMID: 26740381 Review. - International Union of Basic and Clinical Pharmacology CIII: Chemerin Receptors CMKLR1 (Chemerin1) and GPR1 (Chemerin2) Nomenclature, Pharmacology, and Function.
Kennedy AJ, Davenport AP. Kennedy AJ, et al. Pharmacol Rev. 2018 Jan;70(1):174-196. doi: 10.1124/pr.116.013177. Epub 2017 Dec 26. Pharmacol Rev. 2018. PMID: 29279348 Free PMC article. Review.
Cited by
- Chemokine profile in the serum of patients with leptospirosis.
Mariano IHM, Blanco RM, de Souza CE, de Freitas GS, Ho PL, Martins EAL, Romero EC, da Silva JB. Mariano IHM, et al. Front Cell Infect Microbiol. 2024 Oct 29;14:1484291. doi: 10.3389/fcimb.2024.1484291. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39534703 Free PMC article. - Chemokine Receptor N-Terminus Charge Dictates Reliance on Post-Translational Modifications for Effective Ligand Capture and Following Boosting by Defense Peptides.
Xu T, Schou AS, Lackman JJ, Barrio-Calvo M, Verhallen L, Goth CK, Jensen BAH, Veldkamp CT, Volkman BF, Peterson FC, Hjortø GM. Xu T, et al. Int J Mol Sci. 2024 Oct 9;25(19):10854. doi: 10.3390/ijms251910854. Int J Mol Sci. 2024. PMID: 39409188 Free PMC article. - The role of cytokines from salivary gland epithelial cells in the immunopathology of Sjögren's syndrome.
Dong Y, Wang T, Wu H. Dong Y, et al. Front Immunol. 2024 Sep 13;15:1443455. doi: 10.3389/fimmu.2024.1443455. eCollection 2024. Front Immunol. 2024. PMID: 39346911 Free PMC article. Review. - Expression and regulation of the CXCL9-11 chemokines and CXCR3 receptor in Atlantic salmon (Salmo salar).
Valdés N, Espinoza D, Pareja-Barrueto C, Olate N, Barraza-Rojas F, Benavides-Larenas A, Cortés M, Imarai M. Valdés N, et al. Front Immunol. 2024 Sep 5;15:1455457. doi: 10.3389/fimmu.2024.1455457. eCollection 2024. Front Immunol. 2024. PMID: 39301034 Free PMC article. - Development of specific anti-mouse atypical chemokine receptor 4 monoclonal antibodies.
Hirose M, Suzuki H, Ubukata R, Tanaka T, Kaneko MK, Kato Y. Hirose M, et al. Biochem Biophys Rep. 2024 Sep 7;40:101824. doi: 10.1016/j.bbrep.2024.101824. eCollection 2024 Dec. Biochem Biophys Rep. 2024. PMID: 39290345 Free PMC article.
References
- Abel S, Hundhausen C, Mentlein R, Schulte A, Berkhout TA, Broadway N, Hartmann D, Sedlacek R, Dietrich S, Muetze B, et al. (2004) The transmembrane CXC-chemokine ligand 16 is induced by IFN-gamma and TNF-alpha and shed by the activity of the disintegrin-like metalloproteinase ADAM10. J Immunol 172:6362–6372 - PubMed
- Acosta-Rodriguez EV, Rivino L, Geginat J, Jarrossay D, Gattorno M, Lanzavecchia A, Sallusto F, Napolitani G. (2007) Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat Immunol 8:639–646 - PubMed
- Addison CL, Daniel TO, Burdick MD, Liu H, Ehlert JE, Xue YY, Buechi L, Walz A, Richmond A, Strieter RM. (2000) The CXC chemokine receptor 2, CXCR2, is the putative receptor for ELR+ CXC chemokine-induced angiogenic activity. J Immunol 165:5269–5277 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- G0901113/MRC_/Medical Research Council/United Kingdom
- R37 AI040618/AI/NIAID NIH HHS/United States
- G0802838/MRC_/Medical Research Council/United Kingdom
- 099251/Wellcome Trust/United Kingdom
- ImNIH/Intramural NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous