Landscape of genetic lesions in 944 patients with myelodysplastic syndromes - PubMed (original) (raw)
doi: 10.1038/leu.2013.336. Epub 2013 Nov 13.
Y Nagata 2, V Grossmann 1, Y Okuno 3, U Bacher 1, G Nagae 4, S Schnittger 1, M Sanada 2, A Kon 2, T Alpermann 1, K Yoshida 2, A Roller 1, N Nadarajah 1, Y Shiraishi 5, Y Shiozawa 2, K Chiba 5, H Tanaka 6, H P Koeffler 7, H-U Klein 8, M Dugas 8, H Aburatani 4, A Kohlmann 1, S Miyano 9, C Haferlach 1, W Kern 1, S Ogawa 2
Affiliations
- PMID: 24220272
- PMCID: PMC3918868
- DOI: 10.1038/leu.2013.336
Free PMC article
Landscape of genetic lesions in 944 patients with myelodysplastic syndromes
T Haferlach et al. Leukemia. 2014 Feb.
Free PMC article
Abstract
High-throughput DNA sequencing significantly contributed to diagnosis and prognostication in patients with myelodysplastic syndromes (MDS). We determined the biological and prognostic significance of genetic aberrations in MDS. In total, 944 patients with various MDS subtypes were screened for known/putative mutations/deletions in 104 genes using targeted deep sequencing and array-based genomic hybridization. In total, 845/944 patients (89.5%) harbored at least one mutation (median, 3 per patient; range, 0-12). Forty-seven genes were significantly mutated with TET2, SF3B1, ASXL1, SRSF2, DNMT3A, and RUNX1 mutated in >10% of cases. Many mutations were associated with higher risk groups and/or blast elevation. Survival was investigated in 875 patients. By univariate analysis, 25/48 genes (resulting from 47 genes tested significantly plus PRPF8) affected survival (P<0.05). The status of 14 genes combined with conventional factors revealed a novel prognostic model ('Model-1') separating patients into four risk groups ('low', 'intermediate', 'high', 'very high risk') with 3-year survival of 95.2, 69.3, 32.8, and 5.3% (P<0.001). Subsequently, a 'gene-only model' ('Model-2') was constructed based on 14 genes also yielding four significant risk groups (P<0.001). Both models were reproducible in the validation cohort (n=175 patients; P<0.001 each). Thus, large-scale genetic and molecular profiling of multiple target genes is invaluable for subclassification and prognostication in MDS patients.
Figures
Figure 1
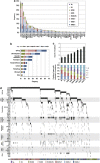
Significantly mutated genes in MDS. (a) Frequency of mutations in 47 significantly mutated genes in 944 cases with different WHO subtypes, which are shown in indicated colors. (b) Frequency of gene mutations involved in common functional pathways, which are defined in Supplementary Table S3. (c) Number of gene mutations detected in different MDS subtypes. (d) Distribution of mutations/deletions of significantly mutated genes in 944 MDS cases.
Figure 2
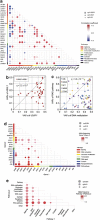
Comparison of mutation loads between major gene targets in MDS. (a) Correlations between major genetic lesions, where the correlation coefficients are indicated by a color gradient and show diagonal plots of variant allele frequencies (VAFs) between ASXL1 and U2AF1 mutations (b) and between mutations in RAS pathway genes (CBL, KRAS, NF1, NRAS and PTPN11) and DNA methylation-related genes (TET2, IDH1/2 and DNMT3A) (c), in which copy number-adjusted VAF values between indicated mutations or mutations of indicated functional pathways were compared using paired T-tests. Comparison was made exhaustively between all possible combinations of commonly mutated genes (>2.5% of mutation rates) (d) or gene pathways (e) with adjustment for multiple testing. Significance (_q_-values) and difference (relative difference in VAFs) is indicated by the size of circles and color gradient, as indicated, respectively.
Figure 3
Illustration of hazard ratios for Model-1 and Model-2. Hazard ratios (HRs, given in numbers) as well as logHR and their 95% confidential intervals (given as chart) for parameters used for Model-1 including clinical and genetic variables (a) and for Model-2 including only genetic variables (referring to the training cohort) (b) are plotted. For a, baseline level for the analysis was the respective IPSS-R risk category with the least risk score (hemoglobin: ⩾10 g/dl, platelets score: ⩾100 × 109/l, blast score: ⩽2%, cytogenetic score very good).
Figure 4
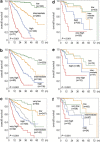
Development of a novel prognostic risk classification. (a) Kaplan–Meier estimates of OS in months (m) for four groups according to Model-1 in the training cohort. Three-year OS for low, intermediate, high and very high-risk groups amounts to 95.2, 69.3, 32.8 and 5.3%, respectively. (b) Kaplan–Meier estimates of OS for four groups according to Model-2 in the training cohort. Three-year OS for low, intermediate, high and very high-risk groups amounts to 83.3, 66.4, 39.7 and 9.5%, respectively. (c) Kaplan–Meier estimates of OS for five groups according to IPSS-R in the training cohort. Three-year OS for very low, low, intermediate, high and very high-risk groups amounts to 88.2, 73.9, 51.9, 45.3 and 19.6%, respectively. (d) Kaplan–Meier estimates of OS for four groups according to Model-1 in the validation cohort. Three-year OS for low, intermediate, high and very high-risk groups amounts to 88.3, 84.4, 55.7 and 22.8%, respectively. (e) Kaplan–Meier estimates of OS for four groups according to Model-2 in the validation cohort. Three-year OS for low, intermediate, high and very high-risk groups amounts to 83.3, 77.0, 64.1 and 33.3%, respectively. (f) Kaplan–Meier estimates of OS for five groups according to IPSS-R in the validation cohort. Three-year OS was 83.3, 93.7, 59.3 and 57.1% for the very low, low, intermediate and high-risk groups. For the very high-risk group, the median OS was 9.2 months, as 3-year OS was not applicable.
Similar articles
- [A novel prognostic model incorporating genetic profiling for myelodysplastic syndromes].
Nagata Y, Ogawa S. Nagata Y, et al. Rinsho Ketsueki. 2017;58(7):776-786. doi: 10.11406/rinketsu.58.776. Rinsho Ketsueki. 2017. PMID: 28781274 Japanese. - Genetic landscape of recurrent ASXL1, U2AF1, SF3B1, SRSF2, and EZH2 mutations in 304 Chinese patients with myelodysplastic syndromes.
Wu L, Song L, Xu L, Chang C, Xu F, Wu D, He Q, Su J, Zhou L, Xiao C, Zhang Z, Zhao Y, Chen S, Li X. Wu L, et al. Tumour Biol. 2016 Apr;37(4):4633-40. doi: 10.1007/s13277-015-4305-2. Epub 2015 Oct 28. Tumour Biol. 2016. PMID: 26508027 - Mutation status and burden can improve prognostic prediction of patients with lower-risk myelodysplastic syndromes.
Jiang L, Luo Y, Zhu S, Wang L, Ma L, Zhang H, Shen C, Yang W, Ren Y, Zhou X, Mei C, Ye L, Xu W, Yang H, Lu C, Jin J, Tong H. Jiang L, et al. Cancer Sci. 2020 Feb;111(2):580-591. doi: 10.1111/cas.14270. Epub 2019 Dec 24. Cancer Sci. 2020. PMID: 31804030 Free PMC article. - Mutations of myelodysplastic syndromes (MDS): An update.
Ganguly BB, Kadam NN. Ganguly BB, et al. Mutat Res Rev Mutat Res. 2016 Jul-Sep;769:47-62. doi: 10.1016/j.mrrev.2016.04.009. Epub 2016 Jun 23. Mutat Res Rev Mutat Res. 2016. PMID: 27543316 Review. - Somatic mutations and epigenetic abnormalities in myelodysplastic syndromes.
Itzykson R, Kosmider O, Fenaux P. Itzykson R, et al. Best Pract Res Clin Haematol. 2013 Dec;26(4):355-64. doi: 10.1016/j.beha.2014.01.001. Epub 2014 Jan 13. Best Pract Res Clin Haematol. 2013. PMID: 24507812 Review.
Cited by
- Current challenges and unmet medical needs in myelodysplastic syndromes.
Platzbecker U, Kubasch AS, Homer-Bouthiette C, Prebet T. Platzbecker U, et al. Leukemia. 2021 Aug;35(8):2182-2198. doi: 10.1038/s41375-021-01265-7. Epub 2021 May 28. Leukemia. 2021. PMID: 34045662 Free PMC article. Review. - Cytotoxic Therapy-Induced Effects on Both Hematopoietic and Marrow Stromal Cells Promotes Therapy-Related Myeloid Neoplasms.
Stoddart A, Wang J, Fernald AA, Davis EM, Johnson CR, Hu C, Cheng JX, McNerney ME, Le Beau MM. Stoddart A, et al. Blood Cancer Discov. 2020 Jul;1(1):32-47. doi: 10.1158/2643-3230.BCD-19-0028. Blood Cancer Discov. 2020. PMID: 32924016 Free PMC article. - Comprehensive Molecular Profiling of Archival Bone Marrow Trephines Using a Commercially Available Leukemia Panel and Semiconductor-Based Targeted Resequencing.
Bartels S, Schipper E, Kreipe HH, Lehmann U. Bartels S, et al. PLoS One. 2015 Jul 29;10(7):e0133930. doi: 10.1371/journal.pone.0133930. eCollection 2015. PLoS One. 2015. PMID: 26222071 Free PMC article. - The genetics of myelodysplastic syndromes and the opportunities for tailored treatments.
Kontandreopoulou CN, Kalopisis K, Viniou NA, Diamantopoulos P. Kontandreopoulou CN, et al. Front Oncol. 2022 Oct 20;12:989483. doi: 10.3389/fonc.2022.989483. eCollection 2022. Front Oncol. 2022. PMID: 36338673 Free PMC article. Review. - Mutational profiling of therapy-related myelodysplastic syndromes and acute myeloid leukemia by next generation sequencing, a comparison with de novo diseases.
Ok CY, Patel KP, Garcia-Manero G, Routbort MJ, Fu B, Tang G, Goswami M, Singh R, Kanagal-Shamanna R, Pierce SA, Young KH, Kantarjian HM, Medeiros LJ, Luthra R, Wang SA. Ok CY, et al. Leuk Res. 2015 Mar;39(3):348-54. doi: 10.1016/j.leukres.2014.12.006. Epub 2014 Dec 20. Leuk Res. 2015. PMID: 25573287 Free PMC article.
References
- Brunning R, Orazi A, Germing U, Le Beau MM, Porwit A, Baumann I, et al. Myelodysplastic syndromesIn: Swerdlow S, Campo E, Lee Harris N, Jaffe ES, Pileri SA, Stein H, et al(eds). 4 edn,WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues 200887–93.
- Cazzola M, la Porta MG, Travaglino E, Malcovati L. Classification and prognostic evaluation of myelodysplastic syndromes. Semin Oncol. 2011;38:627–634. - PubMed
- Greenberg P, Cox C, Le Beau MM, Fenaux P, Morel P, Sanz G, et al. International Scoring System for evaluating prognosis in myelodysplastic syndromes. Blood. 1997;89:2079–2088. - PubMed
- Malcovati L, Germing U, Kuendgen A, Della Porta MG, Pascutto C, Invernizzi R, et al. Time-dependent prognostic scoring system for predicting survival and leukemic evolution in myelodysplastic syndromes. J Clin Oncol. 2007;25:3503–3510. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous