Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes - PubMed (original) (raw)
Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes
Rossella Crescitelli et al. J Extracell Vesicles. 2013.
Abstract
Introduction: In recent years, there has been an exponential increase in the number of studies aiming to understand the biology of exosomes, as well as other extracellular vesicles. However, classification of membrane vesicles and the appropriate protocols for their isolation are still under intense discussion and investigation. When isolating vesicles, it is crucial to use systems that are able to separate them, to avoid cross-contamination.
Method: EVS RELEASED FROM THREE DIFFERENT KINDS OF CELL LINES: HMC-1, TF-1 and BV-2 were isolated using two centrifugation-based protocols. In protocol 1, apoptotic bodies were collected at 2,000×g, followed by filtering the supernatant through 0.8 µm pores and pelleting of microvesicles at 12,200×g. In protocol 2, apoptotic bodies and microvesicles were collected together at 16,500×g, followed by filtering of the supernatant through 0.2 µm pores and pelleting of exosomes at 120,000×g. Extracellular vesicles were analyzed by transmission electron microscopy, flow cytometry and the RNA profiles were investigated using a Bioanalyzer(®).
Results: RNA profiles showed that ribosomal RNA was primary detectable in apoptotic bodies and smaller RNAs without prominent ribosomal RNA peaks in exosomes. In contrast, microvesicles contained little or no RNA except for microvesicles collected from TF-1 cell cultures. The different vesicle pellets showed highly different distribution of size, shape and electron density with typical apoptotic body, microvesicle and exosome characteristics when analyzed by transmission electron microscopy. Flow cytometry revealed the presence of CD63 and CD81 in all vesicles investigated, as well as CD9 except in the TF-1-derived vesicles, as these cells do not express CD9.
Conclusions: Our results demonstrate that centrifugation-based protocols are simple and fast systems to distinguish subpopulations of extracellular vesicles. Different vesicles show different RNA profiles and morphological characteristics, but they are indistinguishable using CD63-coated beads for flow cytometry analysis.
Keywords: RNA; apoptotic bodies; characterization; electron microscopy; exosomes; extracellular vesicles; microvesicles; ultracentrifugation.
Figures
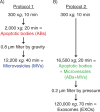
Fig. 1
Flow chart over two different differential centrifugation-based protocols. Apoptotic bodies (ABs) and microvesicles (MVs) were isolated separately using protocol 1 (A). ABs and MVs were isolated together (ABs+MVs) followed by exosome (EXO) isolation, using protocol 2 (B).
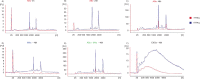
Fig. 2
RNA profiles from different subpopulations of extracellular vesicles (EVs). RNA was extracted from vesicles released by three different cell lines; HMC-1 (human mast cell line), TF-1 (human erythroleukemia cells) and BV-2 (mouse microglia cells). The electropherograms show the size distribution in nucleotides (nt) and fluorescence intensity (FU) of total RNA in apoptotic bodies (ABs), microvesicles (MVs), ABs and MVs together (ABs+MVs) and exosomes (EXOs). The short peak at 25 nt is an internal standard. (A) In ABs the most dominant peaks are the 18S and 28S ribosomal RNA. (B) The 18S and 28S rRNA are not evident in MVs from HMC-1 and BV-2, but only obvious in MVs from TF-1, however at low concentrations. (C) 18S and 28S peaks are evident in the pellet composed by ABs and MVs together (ABs+MVs). (D) In EXOs small RNA is dominating, with no or very small rRNA peaks detected. (E–G) The overlapping profiles from ABs (in red) and MVs (in blue) and both collected together (ABs+MVs − in green), suggesting that the contribution of 18S and 28S rRNA is by ABs. The electropherograms are representative of n=4.
Fig. 3
RNA profiles from different subpopulations of extracellular vesicles (EVs) after TRAIL-induced apoptosis. The electropherograms show the RNA size distribution in nucleotides (nt) and fluorescence intensity (FU) in apoptotic bodies (ABs), microvesicles (MVs), ABs and MVs together (ABs+MVs) and exosomes (EXOs) in TF-1 cells with and without TRAIL treatment. The short peak at 25 nt is an internal standard. (A–C) RNA profiles from ABs released by TF-1 cells after 4, 24, 48 hours of TRAIL treatment (in blue) and without TRAIL (in red). After 4 hours (A), 24 hours (B) and 48 hours (C) of TRAIL treatment, in ABs, the peaks of 18S and 28S rRNAs are more prominent comparing with ABs released in the absence of TRAIL. (D–F) RNA profiles from MVs, ABs+MVs and EXOs released by TF-1 cells after 48 hours of TRAIL treatment (in blue) and without TRAIL (in red). (D) The low 18S and 28S rRNA peaks in MVs without TRAIL (in red) become much more prominent after TRAIL treatment (in blue). (E) The highest rRNA peaks are seen in the pellet composed by ABs and MVs together (ABs+MVs). (F) After 48 hours of TRAIL-induced apoptosis, increased amount of small RNAs is observed in exosomes (EXOs). The electropherograms are representative of n=2.
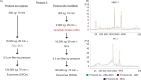
Fig. 4
Flow chart over the original and modified protocol 2. (A) In the modification of protocol 2, a 2,000×g step was added to isolate apoptotic bodies (ABs) and microvesicles (MVs) separately, prior to EXOs isolation (here called protocol 2b). (B) The RNA profiles from the different subpopulation of extracellular vesicles (EVs) collected using protocol 2a and 2b. RNA was extracted from vesicles releases from two different cell lines; HMC-1 and TF-1. Shown here are the overlapping profiles from ABs (ABs – in red), MVs (MVs – in blue) and both of them collected together (ABs+MVs − in green), indicating that the contribution of 18S and 28S rRNA is primarily by ABs. The electopherograms show the size distribution in nucleotides (nt) and fluorescence intensity (FU) of total RNA. The peak at 25 nt is an internal standard. The electropherograms are representative of n=3.
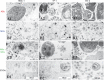
Fig. 5
Analysis of ABs, MVs and EXOs by TEM. Micrographs of vesicles released from three different cell lines; HMC-1 (human mast cell line), TF-1 (human erythroleukemia cells), and BV-2 (mouse microglia cells) are shown. (A1–3) Dense structures show the chromatin substance in the generally round shaped apoptotic bodies (ABs) with a size of 800–5,000 nm. (B1–3) Microvesicles (MVs) are diverse in their shape and density, with a size range between 200 and 800 nm. (C1–3) In the pellet obtained by centrifugation at 16,500×g presents the mixture of ABs and MVs. (D1–3) The exosome (EXO) fraction from HMC-1 (D1), TF-1 (D2) and BV-2 (D3) cells were found to have a diameter of approximately 40–100 nm.
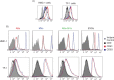
Fig. 6
Detection and characterization of extracellular vesicles (EVs) by flow cytometry. The CD9, CD63 and CD81 expression on HMC-1 and TF-1 cells (A) and their expression on different vesicles, using anti-CD63-coated beads, are shown. (B) Cells and vesicles were immunostained against the tetraspanin (open curve) CD9 (in black), CD63 (in blue) and CD81 (in red) and compared with their appropriate isotype control (filled curve). The graphs are representative of n=3.
Similar articles
- Nanoscale flow cytometry to distinguish subpopulations of prostate extracellular vesicles in patient plasma.
Padda RS, Deng FK, Brett SI, Biggs CN, Durfee PN, Brinker CJ, Williams KC, Leong HS. Padda RS, et al. Prostate. 2019 May;79(6):592-603. doi: 10.1002/pros.23764. Epub 2019 Jan 24. Prostate. 2019. PMID: 30680751 - Exosome-like vesicles in uterine aspirates: a comparison of ultracentrifugation-based isolation protocols.
Campoy I, Lanau L, Altadill T, Sequeiros T, Cabrera S, Cubo-Abert M, Pérez-Benavente A, Garcia A, Borrós S, Santamaria A, Ponce J, Matias-Guiu X, Reventós J, Gil-Moreno A, Rigau M, Colas E. Campoy I, et al. J Transl Med. 2016 Jun 18;14(1):180. doi: 10.1186/s12967-016-0935-4. J Transl Med. 2016. PMID: 27317346 Free PMC article. - Tetraspanins distinguish separate extracellular vesicle subpopulations in human serum and plasma - Contributions of platelet extracellular vesicles in plasma samples.
Karimi N, Dalirfardouei R, Dias T, Lötvall J, Lässer C. Karimi N, et al. J Extracell Vesicles. 2022 May;11(5):e12213. doi: 10.1002/jev2.12213. J Extracell Vesicles. 2022. PMID: 35524458 Free PMC article. - Extracellular vesicles from activated platelets: a semiquantitative cryo-electron microscopy and immuno-gold labeling study.
Brisson AR, Tan S, Linares R, Gounou C, Arraud N. Brisson AR, et al. Platelets. 2017 May;28(3):263-271. doi: 10.1080/09537104.2016.1268255. Epub 2017 Jan 19. Platelets. 2017. PMID: 28102751 Review. - Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies.
Akers JC, Gonda D, Kim R, Carter BS, Chen CC. Akers JC, et al. J Neurooncol. 2013 May;113(1):1-11. doi: 10.1007/s11060-013-1084-8. Epub 2013 Mar 2. J Neurooncol. 2013. PMID: 23456661 Free PMC article. Review.
Cited by
- Glutamine and serum starvation alters the ATP production, oxidative stress, and abundance of mitochondrial RNAs in extracellular vesicles produced by cancer cells.
Bugajova M, Raudenska M, Hanelova K, Navratil J, Gumulec J, Petrlak F, Vicar T, Hrachovinova S, Masarik M, Kalfert D, Grega M, Plzak J, Betka J, Balvan J. Bugajova M, et al. Sci Rep. 2024 Oct 28;14(1):25815. doi: 10.1038/s41598-024-73943-2. Sci Rep. 2024. PMID: 39468126 Free PMC article. - Urinary extracellular vesicles for RNA extraction: optimization of a protocol devoid of prokaryote contamination.
Tataruch-Weinert D, Musante L, Kretz O, Holthofer H. Tataruch-Weinert D, et al. J Extracell Vesicles. 2016 Jun 24;5:30281. doi: 10.3402/jev.v5.30281. eCollection 2016. J Extracell Vesicles. 2016. PMID: 27345058 Free PMC article. - Non-coding RNAs in Exosomes: New Players in Cancer Biology.
Silva M, Melo SA. Silva M, et al. Curr Genomics. 2015 Oct;16(5):295-303. doi: 10.2174/1389202916666150707154719. Curr Genomics. 2015. PMID: 27047249 Free PMC article. - Extracellular Vesicles and Cell-Cell Communication: New Insights and New Therapeutic Strategies Not Only in Oncology.
Gieseler F, Ender F. Gieseler F, et al. Int J Mol Sci. 2020 Jun 18;21(12):4331. doi: 10.3390/ijms21124331. Int J Mol Sci. 2020. PMID: 32570703 Free PMC article. - From Genes to Therapy: Pituitary Adenomas in the Era of Precision Medicine.
Toader C, Dobrin N, Tataru CI, Covache-Busuioc RA, Bratu BG, Glavan LA, Costin HP, Corlatescu AD, Dumitrascu DI, Ciurea AV. Toader C, et al. Biomedicines. 2023 Dec 21;12(1):23. doi: 10.3390/biomedicines12010023. Biomedicines. 2023. PMID: 38275385 Free PMC article. Review.
References
- Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–93. - PubMed
- Keller S, Sanderson MP, Stoeck A, Altevogt P. Exosomes: from biogenesis and secretion to biological function. Immunol Lett. 2006;107:102–8. - PubMed
- Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907–20. - PubMed
- Ratajczak J, Wysoczynski M, Hayek F, Janowska-Wieczorek A, Ratajczak MZ. Membrane-derived microvesicles: important and underappreciated mediators of cell-to-cell communication. Leukemia. 2006;20:1487–95. - PubMed
- Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19:43–51. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous