Molecular basis for DNA double-strand break annealing and primer extension by an NHEJ DNA polymerase - PubMed (original) (raw)
Molecular basis for DNA double-strand break annealing and primer extension by an NHEJ DNA polymerase
Nigel C Brissett et al. Cell Rep. 2013.
Abstract
Nonhomologous end-joining (NHEJ) is one of the major DNA double-strand break (DSB) repair pathways. The mechanisms by which breaks are competently brought together and extended during NHEJ is poorly understood. As polymerases extend DNA in a 5'-3' direction by nucleotide addition to a primer, it is unclear how NHEJ polymerases fill in break termini containing 3' overhangs that lack a primer strand. Here, we describe, at the molecular level, how prokaryotic NHEJ polymerases configure a primer-template substrate by annealing the 3' overhanging strands from opposing breaks, forming a gapped intermediate that can be extended in trans. We identify structural elements that facilitate docking of the 3' ends in the active sites of adjacent polymerases and reveal how the termini act as primers for extension of the annealed break, thus explaining how such DSBs are extended in trans. This study clarifies how polymerases couple break-synapsis to catalysis, providing a molecular mechanism to explain how primer extension is achieved on DNA breaks.
Copyright © 2013 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
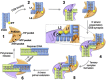
Graphical abstract
Figure 1
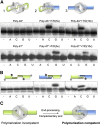
A Functional NHEJ Polymerase-Mediated Synapsis (A) NHEJ reactions were performed with PolDom (600 nM) using a homopolymeric single-stranded DNA substrate (poly-dA or poly-dT) and a 3′-protruding substrate formed with the oligonucleotides TTTG or AAAC and NHEJ-D. In this and the other figures, the black spheres indicate the presence of a 5′-P group in the substrate and the star denotes the position of the radioactive label. When indicated, each of the four NTPs (100 μM) were added in the presence of 1 mM MnCl2. (B) NHEJ reactions were performed with PolDom (600 nM) using DNA substrates formed with the oligonucleotides TTTG with NHEJ-D and AAAC with NHEJ-D2. When indicated, each of the four NTPs (100 μM) was added in the presence of 1 mM MnCl2. (C) A stable dimeric complex formed at noncomplementary DNA ends (polymerization incompetent) would allow further nucleolytic resection to produce a polymerization competent DNA substrate. See also Figure S1.
Figure 2
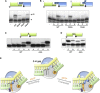
Architecture of an Annealed dsDNA Break Bound to an NHEJ Polymerase Schematic representation of the annealed DNA double-strand break present in the crystal structure with the annealed microhomology sequence highlighted. Below this scheme are representations of the crystal structure of the annealed DNA double-strand break bound to an NHEJ polymerase, PolDom. The figure depicts a synaptic complex formed between two binary (DNA [T/D]-PolDom) complexes that have come together, in a “face-to-face” orientation, by annealing of the 3′ self-complementary DNA overhangs of the break. To the left of the figure, the polymerase is depicted as a gray solvent accessible surface, and the DNA is depicted in red or green (side-on and top-down views). The polymerases facilitate DNA break synapsis between discontinuous DNA ends by cradling the termini, within a continuous molecular surface, promoting microhomology-mediated end synapsis. The middle of the figure has a protein ribbon representation of the structure of the annealed DNA break bound to PolDom (side-on and top-down views). The polymerase monomers are colored light blue and yellow, respectively. Significant structural elements loop 1 and loop 2 are colored blue and cyan, respectively. The polymerase induces a major splaying (∼119°) of the template strand (T). The resulting 3′ overhangs are annealed together, forming four Watson-Crick base pairs (G7-C10), via a region of microhomology. This end synapsis is promoted by interactions with loops 1 and 2 (inset). The template strand from one binary complex terminates in the active site of the opposing binary complex, effectively becoming an incoming primer strand (inset). The inset also features catalytic site residues (tan) as well as residues involved in template strand splaying and orientation (yellow) and primer strand orientation and tethering (cyan) (see Figures 3A, 6A, 6B, 7A, 7B, and S4–S7 for more detail). See also Figure S2.
Figure 3
Formation of Functional NHEJ Complexes on Short Overhangs: Role of 5′ Phosphate Binding and Dimeric versus Monomeric Configurations (A) Schematic representation of the phosphate binding region and DNA ds/ss (T/D) junction of annealed break DNA bound to PolDom. The protein is depicted as a translucent solvent accessible surface and DNA (red) is depicted with protein side-chain neighbors that are within 4 Å of the strand (blue). The 5′-phosphate is depicted as scaled van der Waals spheres and the phosphate atom (purple) is bound in a pocket formed by conserved residues (Asn13, Lys16, and Lys26). DNA at the ds/ss junction is wedged against Arg53 and Pro55, and the template strand (T) splayed out by Phe63 and Phe64 with the templating base (yellow) interacting with Phe64. (B) NHEJ reactions were performed with PolDom (600 nM) using various substrates formed with the oligonucleotides TTG with NHEJ-D and AAC with NHEJ-D2. When indicated, each of the four NTPs (100 μM) were added in the presence of 1 mM MnCl2. (C) Footprinting assays with Polβ (5 μg) or PolDom (5 μg) were conducted as described in Experimental Procedures. BSA (10 μg) was added to the control lane. The substrate was formed with oligonucleotides FP-T, FP-P, and FP-D, depicted on the right. See also Figure S3.
Figure 4
Residues Contacting the Template Strand: Implications for PolDom-Mediated NHEJ Reactions (A) EMSAs were performed for the indicated proteins (200 nM) using a gapped substrate containing the oligonucleotides SP1C, T13C, and DG-P. When indicated, 1 mM MnCl2 and/or 100 μM UTP was added. After electrophoresis, the gel was dried and the labeled fragments were detected by autoradiography. (B) Footprinting assays of wild-type or mutant PolDom (5 μg) were conducted as described in the Experimental Procedures. BSA (10 μg) was added to the control lane. The substrate was formed with oligonucleotides FP-T, FP-P, and FP-D, depicted on the right. (C) Gap-filling reactions were performed as described in Experimental Procedures for the indicated proteins (25 nM) using a gapped DNA substrate containing the oligonucleotides SP1C, T13C, and DG-P. When indicated, NTPs were added separately at 10 nM in the presence of 1 mM MnCl2. (D) NHEJ reactions were performed with 600 nM of the indicated proteins using a set of DNA substrates formed with the oligonucleotides TG with NHEJ-D and AC with NHEJ-D2. When indicated, each of the four NTPs (100 μM) was added in the presence of 1 mM MnCl2. See also Figure S4.
Figure 5
Selecting the Templating Base: Roles of Residues Phe63 and Phe64 (A and B) NHEJ reactions were performed with 600 nM of the indicated proteins using a set of DNA substrates formed by hybridizing the oligonucleotides CCG with NHEJ-D and GGC with NHEJ-D2. In (A), only GTP (100 μM) was added in the presence of 1 mM MnCl2, whereas in (B) the other three nucleotides were added (100 μM). (C and D) Gap-filling reactions were performed as described in Experimental Procedures for the indicated proteins (25 nM) using a gapped DNA substrate containing the oligonucleotides SP1C, T13C, and DG-P (C) or P15, T17, and DG2P (D). When indicated, NTPs were added separately at 10 nM in the presence of 1 mM MnCl2. (E) A cartoon showing the dichotomy that PolDom confronts when dealing with gaps longer than 1 nt during NHEJ; the template strand is either “scrunched,” and the gap filled in correctly (left side), or the template strand is dislocated and sequence is lost with the production of frameshifts (right side). The protein is shown as a gray surface with a blue section indicating the approximate position of loop 1, 5′P and incoming nucleotide are colored orange, the two metal ions are shown in purple, and the DNA substrate is shown in yellow (template strand) and green (primer and downstream strands). Phenylalanines Phe63 and Phe64 are shown as blue hexagons holding the kink in the DNA substrate, indicating with a darker blue color their importance for each reaction. See main text for details.
Figure 6
Transition of the Template Strand to Incoming Primer Strand in the Gapped Complex (A) Interaction of loop 1 residues (blue) and their involvement in directing the templating DNA strand (red). A translucent gray surface further depicts the protein solvent accessible surface. His83, Arg84, and Ser85 of loop 1 directs the template DNA that has been splayed by Phe63 and Phe63 toward the opposing protein monomer. (B) Rotated view of (A) showing the incoming primer strand (red) as it accepted in trans into the active site of the opposite protein monomer. Loop 2 and the major interacting residues are colored cyan. A translucent gray solvent accessible surface further depicts the opposing protein monomer. Conserved residues Met215, Lys217, and Arg220 contact the incoming primer as the 3′-OH is stabilized by contacts in the active site (green). (C) NHEJ reactions were performed with 600 nM PolDom using various DNA substrates formed with the oligonucleotides TTTG, TTG, or TG with NHEJ-D and AAAC, AAC, or AC with NHEJ-D2. When indicated, each of the four NTPs (100 μM) were added in the presence of 1 mM MnCl2. (D) EMSA assays were performed for the indicated proteins (200 nM) using a gapped DNA substrate containing the oligonucleotides SP1C, T13C, and DG-P. (E) Gap-filling reactions were performed as described in Experimental Procedures for the indicated proteins (25 nM) using a gapped DNA substrate containing the oligonucleotides SP1C, T13C, and DG-P. When indicated, NTPs were added separately at 10 nM in the presence of 1 mM MnCl2. See also Figures S5 and S6.
Figure 7
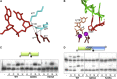
In trans Docking of a Primer Terminus in the Polymerase Active Site (A) Interaction and stabilization of the 3′-hydroxyl (3′-OH) of the incoming primer within the active site of PolDom. Residues Lys235, Ser229, and Gln230 (cyan) form a network that interacts with the 3′-OH terminus of the primer strand (red). Two of the catalytic aspartates Asp137 and Asp227 (brown) are also part of this network. (B) A UTP molecule (tan) and catalytic metal ions (magenta), from a PolDom preternary structure (PDB:
3PKY
), were superposed into the active site of the annealed break DNA-PolDom complex. The 3′-OH terminus of the primer strand (red) is within nucleophilic attacking distance of the α-phosphate of UTP, providing compelling evidence that this represents a preternary in trans configuration awaiting the arrival of metal ions an incoming base. The templating DNA strand is depicted (green), and the catalytic residues are colored as in (A). (C) Gap-filling reactions were performed as described in Experimental Procedures for the indicated proteins (25 nM) using a gapped DNA substrate containing the oligonucleotides SP1C, T13C, and DG-P. When indicated, NTPs were added separately at 10 nM in the presence of 1 mM MnCl2. (D) NHEJ reactions were performed with 600 nM PolDom using a set of DNA substrates formed with the oligonucleotides D3 and NHEJ-D (green, fast running species on the gel) or D4 and NHEJ-D2 (blue, slow running species). Both oligonucleotides were labeled so that primer extension can be observed on both sides of the break at the same time. As indicated, each of the four NTPs (100 μM) was added in the presence of 1 mM MnCl2. See also Figure S7.
Similar articles
- Ribonucleolytic resection is required for repair of strand displaced nonhomologous end-joining intermediates.
Bartlett EJ, Brissett NC, Doherty AJ. Bartlett EJ, et al. Proc Natl Acad Sci U S A. 2013 May 28;110(22):E1984-91. doi: 10.1073/pnas.1302616110. Epub 2013 May 13. Proc Natl Acad Sci U S A. 2013. PMID: 23671117 Free PMC article. - Nonhomologous DNA end-joining for repair of DNA double-strand breaks.
Pannunzio NR, Watanabe G, Lieber MR. Pannunzio NR, et al. J Biol Chem. 2018 Jul 6;293(27):10512-10523. doi: 10.1074/jbc.TM117.000374. Epub 2017 Dec 14. J Biol Chem. 2018. PMID: 29247009 Free PMC article. Review. - Structural evidence for an in trans base selection mechanism involving Loop1 in polymerase μ at an NHEJ double-strand break junction.
Loc'h J, Gerodimos CA, Rosario S, Tekpinar M, Lieber MR, Delarue M. Loc'h J, et al. J Biol Chem. 2019 Jul 5;294(27):10579-10595. doi: 10.1074/jbc.RA119.008739. Epub 2019 May 28. J Biol Chem. 2019. PMID: 31138645 Free PMC article. - Analysis of chromatid-break-repair detects a homologous recombination to non-homologous end-joining switch with increasing load of DNA double-strand breaks.
Murmann-Konda T, Soni A, Stuschke M, Iliakis G. Murmann-Konda T, et al. Mutat Res Genet Toxicol Environ Mutagen. 2021 Jul;867:503372. doi: 10.1016/j.mrgentox.2021.503372. Epub 2021 Jun 12. Mutat Res Genet Toxicol Environ Mutagen. 2021. PMID: 34266628 - The mechanism of human nonhomologous DNA end joining.
Lieber MR. Lieber MR. J Biol Chem. 2008 Jan 4;283(1):1-5. doi: 10.1074/jbc.R700039200. Epub 2007 Nov 12. J Biol Chem. 2008. PMID: 17999957 Review.
Cited by
- Plant organellar DNA polymerases repair double-stranded breaks by microhomology-mediated end-joining.
García-Medel PL, Baruch-Torres N, Peralta-Castro A, Trasviña-Arenas CH, Torres-Larios A, Brieba LG. García-Medel PL, et al. Nucleic Acids Res. 2019 Apr 8;47(6):3028-3044. doi: 10.1093/nar/gkz039. Nucleic Acids Res. 2019. PMID: 30698803 Free PMC article. - Bacterial Ligase D preternary-precatalytic complex performs efficient abasic sites processing at double strand breaks during nonhomologous end joining.
de Ory A, Carabaña C, de Vega M. de Ory A, et al. Nucleic Acids Res. 2019 Jun 4;47(10):5276-5292. doi: 10.1093/nar/gkz265. Nucleic Acids Res. 2019. PMID: 30976810 Free PMC article. - Molecular basis of microhomology-mediated end-joining by purified full-length Polθ.
Black SJ, Ozdemir AY, Kashkina E, Kent T, Rusanov T, Ristic D, Shin Y, Suma A, Hoang T, Chandramouly G, Siddique LA, Borisonnik N, Sullivan-Reed K, Mallon JS, Skorski T, Carnevale V, Murakami KS, Wyman C, Pomerantz RT. Black SJ, et al. Nat Commun. 2019 Sep 27;10(1):4423. doi: 10.1038/s41467-019-12272-9. Nat Commun. 2019. PMID: 31562312 Free PMC article. - Structural Determinants Responsible for the Preferential Insertion of Ribonucleotides by Bacterial NHEJ PolDom.
Sánchez-Salvador A, de Vega M. Sánchez-Salvador A, et al. Biomolecules. 2020 Jan 30;10(2):203. doi: 10.3390/biom10020203. Biomolecules. 2020. PMID: 32019147 Free PMC article. - Stringent Primer Termination by an Archaeo-Eukaryotic DNA Primase.
Bergsch J, Devillier JC, Jeschke G, Lipps G. Bergsch J, et al. Front Microbiol. 2021 Apr 13;12:652928. doi: 10.3389/fmicb.2021.652928. eCollection 2021. Front Microbiol. 2021. PMID: 33927705 Free PMC article.
References
- Brissett N.C., Pitcher R.S., Juarez R., Picher A.J., Green A.J., Dafforn T.R., Fox G.C., Blanco L., Doherty A.J. Structure of a NHEJ polymerase-mediated DNA synaptic complex. Science. 2007;318:456–459. - PubMed
- Brissett N.C., Martin M.J., Pitcher R.S., Bianchi J., Juarez R., Green A.J., Fox G.C., Blanco L., Doherty A.J. Structure of a preternary complex involving a prokaryotic NHEJ DNA polymerase. Mol. Cell. 2011;41:221–231. - PubMed
- Chapman J.R., Taylor M.R., Boulton S.J. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell. 2012;47:497–510. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials