Gle1 functions during mRNA export in an oligomeric complex that is altered in human disease - PubMed (original) (raw)
Gle1 functions during mRNA export in an oligomeric complex that is altered in human disease
Andrew W Folkmann et al. Cell. 2013.
Abstract
The conserved multifunctional protein Gle1 regulates gene expression at multiple steps: nuclear mRNA export, translation initiation, and translation termination. A GLE1 mutation (FinMajor) is causally linked to human lethal congenital contracture syndrome-1 (LCCS1); however, the resulting perturbations on Gle1 molecular function were unknown. FinMajor results in a proline-phenylalanine-glutamine peptide insertion within the uncharacterized Gle1 coiled-coil domain. Here, we find that Gle1 self-associates both in vitro and in living cells via the coiled-coil domain. Electron microscopy reveals that high-molecular-mass Gle1 oligomers form ?26 nm diameter disk-shaped particles. With the Gle1-FinMajor protein, these particles are malformed. Moreover, functional assays document a specific requirement for proper Gle1 oligomerization during mRNA export, but not for Gle1's roles in translation. These results identify a mechanistic step in Gle1's mRNA export function at nuclear pore complexes and directly implicate altered export in LCCS1 disease pathology.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
Figure 1. Conserved structural and functional elements of Gle1 from S. cerevisiae and humans
(A) Diagram depicting functional and structural domains in S. cerevisiae (y)Gle1. Black arrow represents the position of the y-gle1-4 mutation (G382R) (Watkins et al., 1998). White arrow marks the location of conserved IP6-coordinating residues K377 and K378 (Alcazar-Roman et al., 2010). (B) Diagram depicting functional and structural domains in human (h)Gle1B (adapted from Kendirgi et al., 2005). “PFQ” denotes location of the FinMajor insertion after amino acid 144 in the predicted coiled-coil domain (Nousiainen et al., 2008). Black arrow represents the homologous position (Q548) of the residue mutated in y-gle1-4 (Watkins et al., 1998). White arrow marks the location of conserved IP6-coordinating residues K526 and K527 (Alcazar-Roman et al., 2010). (C) Paircoil2-generated structure predictions of Gle1 polypeptide sequences, showing the structural effect of h-gle1-FinMajor and y-gle1 engineered PFQ insertions. (+) indicates >50% probability of coiled-coil structure. PFQ insertions locations are designated by bold underlined typeface. See Table S1 and S2 for Paircoil2 scores and p-values.
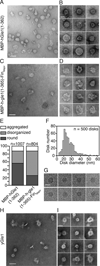
Figure 2. Gle1 oligomers form disk structures
(A–B) hGle1 forms large circular structures. (A) Representative EM image for purified MBP-hGle1(1–362). Bar, 50 nm. (B) Gallery of individual MBP-hGle1(1–362) particles. Bar, 25 nm. (C–E) FinMajor particles are malformed and disorganized. (C) Representative EM image of MBP-h-gle1(1–365)-FinMajor. Bar, 50 nm. (D) Gallery of individual MBP-h-gle1(1–365)-FinMajor particles. Bar, 25 nm. (E) Quantification of particle morphology for MBP-hGle1(1–362) and MBP-h-gle1(1–365)-FinMajor samples, categorized as aggregates, disorganized, or round. (F) hGle1 particles vary in diameter. Histogram of the measured diameter of MBP-hGle1(1–362) particles. (G) hGle1 oligomeric particles form disk-like structures. CryoEM images of MBP-hGle1(1–362) disk-shaped structures in vitrified ice. Bar, 25 nm. (H–I) Oligomeric disk structures are conserved through evolution. (H) Representative EM image of recombinant yGle1. Bar, 50 nm. (I) Gallery of individual yGle1 particles. Bar, 25 nm.
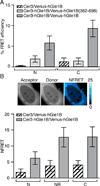
Figure 3. hGle1B self-associates in living cells
(A) hGle1B self-associates in living cells. Analysis of hGle1B interactions by acceptor photobleaching FRET microscopy in HeLa cells expressing the indicated fluorescent protein FRET pairs. FRET efficiencies of indicated regions were measured. Nucleoplasm and cytoplasm are designated by “N” and “C”, respectively. Error bars represent mean ± 95% confidence interval (CI) with n ≥ 20 cells from two independent experiments. (B) hGle1B self-associates at the nuclear rim. Analysis of sensitized emission FRET at the nuclear rim in HeLa cells expressing the indicated proteins. Shown are representative images of Venus-hGle1B (acceptor), Cer-hGle1B (donor) and the normalized FRET (NFRET) intensity map signal, top. Bar, 10µm. Bar graph depicts the NFRET signal for indicated regions, bottom. Nucleoplasm, cytoplasm, and nuclear rim are abbreviated as “N”,”C”, and “NR”, respectively. Error bars for each condition represent mean ± 95% CI with n ≥ 15 cells from at least two independent experiments.
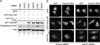
Figure 4. FinMajor has a defect in nuclear poly(A)+ RNA export
(A) hGLE1 siRNA treatment depletes endogenous hGle1 protein levels. Immunoblot analysis of hGle1 and actin protein levels in scrambled control (CTRL) or hGLE1 siRNA-treated HeLa cells transfected with the indicated GFP-tagged proteins. (B) Nuclear poly(A)+ RNA accumulation in hGLE1 siRNA treated cells expressing the indicated GFP-tagged proteins, detected by in situ oligo-dT hybridization and direct fluorescence microscopy. See Figure S3B for quantification of the nucleocytoplasmic distribution of poly(A)+ RNA.
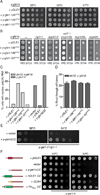
Figure 5. The FinMajor mimic y-gle1^PFQ alleles have specific defects in mRNA export
(A) y-gle1^PFQ mutants exhibit no growth defect. Growth of the indicated strains in 5-fold serial dilution on YPD was monitored at the temperatures shown.(B) y-gle1^PFQ_mutants display genetic interactions with mRNA export mutants. Strains bearing the indicated mutation in combination with y-gle1Δ harboring a y-gle1^PFQ-LEU plasmid and a yGLE1/URA3 plasmid were monitored for growth at 23°C. Failure to grow on synthetic complete media containing 5-FOA indicates synthetic lethality. (C) The yGle1 PFQ insertions that mimic FinMajor perturb mRNA export, an_d y-gle1ΔCC expression does not rescue export defects. Nuclear accumulation of poly(A)+ RNA was detected by in situ oligo-dT hybridization following a shift to 37°C for 2 hours (ipk1Δy-gle1Δ) or 1 hour (y-gle1-4). Calculations were based on >100 cells/condition. See Figure S4D for representative FISH images. (D) y-gle1^PFQ mutants exhibit no defect in translation termination. Ratios of luciferase and β-galactosidase activities were determined and read-through efficiency expressed as the percentage from the reporter with a stop codon inserted in-frame into the linker region between the tandem β-galactosidase and luciferase coding sequences (denoted the TMV reporter) compared to the reporter lacking a stop codon (the TQ control) (Stahl et al., 1995). Standard error of the mean was calculated from 3 independent experiments.(E) y-gle1ΔCC rescues the temperature sensitivity of y-gle1-2 nip1-1. Growth of serially diluted strains on -LEU media was monitored at the indicated temperatures.(F) Oligomerization of Gle1 is required for function in vivo. Mutant y-gle1-4 strains harboring plasmids expressing yGLE1, vector only, y-gle1ΔCC, y-gle1ΔCC+hGLE1-CC, y-gle1ΔCC+GCN4, or y-gle1ΔCC+FinMajor-CC were monitored for growth in 5-fold serial dilution on -LEU media at the temperatures shown. (Left) Schematic representation of the coiled-coil chimeric proteins with yGle1 (red), GCN4 (yellow), hGLE1 (Green), and FinMajor (Green with white bar indicating PFQ insertion).
Figure 6. Nucleocytoplasmic shuttling dynamics are altered for the LCCS1 FinMajor disease protein
(A–B) FinMajor has altered nuclear shuttling activity. (A) FRAP analysis of HeLa cells expressing GFP-hGLE1B and GFP-FinMajor. (B) FRAP analysis of hGLE1 siRNA-treated HeLa cells expressing siRNA-resistant GFP-hGle1BR or GFP-FinMajorR. Bar, 10µm. (C–D) Recovery curves of the experimentally determined bleached region, fit with a one-phase association model. Error bars represent mean ± 95% CI with n ≥ 12 cells from 3 independent experiments.
Similar articles
- Insights into mRNA export-linked molecular mechanisms of human disease through a Gle1 structure-function analysis.
Folkmann AW, Dawson TR, Wente SR. Folkmann AW, et al. Adv Biol Regul. 2014 Jan;54:74-91. doi: 10.1016/j.jbior.2013.10.002. Epub 2013 Nov 13. Adv Biol Regul. 2014. PMID: 24275432 Free PMC article. Review. - Functions of Gle1 are governed by two distinct modes of self-association.
Mason AC, Wente SR. Mason AC, et al. J Biol Chem. 2020 Dec 4;295(49):16813-16825. doi: 10.1074/jbc.RA120.015715. Epub 2020 Sep 27. J Biol Chem. 2020. PMID: 32981894 Free PMC article. - Nup42 and IP6 coordinate Gle1 stimulation of Dbp5/DDX19B for mRNA export in yeast and human cells.
Adams RL, Mason AC, Glass L, Aditi, Wente SR. Adams RL, et al. Traffic. 2017 Dec;18(12):776-790. doi: 10.1111/tra.12526. Epub 2017 Oct 16. Traffic. 2017. PMID: 28869701 Free PMC article. - Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export.
Weirich CS, Erzberger JP, Flick JS, Berger JM, Thorner J, Weis K. Weirich CS, et al. Nat Cell Biol. 2006 Jul;8(7):668-76. doi: 10.1038/ncb1424. Epub 2006 Jun 18. Nat Cell Biol. 2006. PMID: 16783364 - Dbp5 - from nuclear export to translation.
Tieg B, Krebber H. Tieg B, et al. Biochim Biophys Acta. 2013 Aug;1829(8):791-8. doi: 10.1016/j.bbagrm.2012.10.010. Epub 2012 Nov 2. Biochim Biophys Acta. 2013. PMID: 23128325 Review.
Cited by
- The Structure of the Nuclear Pore Complex (An Update).
Lin DH, Hoelz A. Lin DH, et al. Annu Rev Biochem. 2019 Jun 20;88:725-783. doi: 10.1146/annurev-biochem-062917-011901. Epub 2019 Mar 18. Annu Rev Biochem. 2019. PMID: 30883195 Free PMC article. Review. - Probing nuclear pore complex architecture with proximity-dependent biotinylation.
Kim DI, Birendra KC, Zhu W, Motamedchaboki K, Doye V, Roux KJ. Kim DI, et al. Proc Natl Acad Sci U S A. 2014 Jun 17;111(24):E2453-61. doi: 10.1073/pnas.1406459111. Epub 2014 Jun 3. Proc Natl Acad Sci U S A. 2014. PMID: 24927568 Free PMC article. - Global carrier frequency and predicted genetic prevalence of patients with pathogenic sequence variants in autosomal recessive genetic neuromuscular diseases.
Choi WJ, Kim SH, Lee SR, Oh SH, Kim SW, Shin HY, Park HJ. Choi WJ, et al. Sci Rep. 2024 Feb 15;14(1):3806. doi: 10.1038/s41598-024-54413-1. Sci Rep. 2024. PMID: 38361118 Free PMC article. - Regulation of mRNA trafficking by nuclear pore complexes.
Bonnet A, Palancade B. Bonnet A, et al. Genes (Basel). 2014 Sep 2;5(3):767-91. doi: 10.3390/genes5030767. Genes (Basel). 2014. PMID: 25184662 Free PMC article. Review. - Macromolecular transport between the nucleus and the cytoplasm: Advances in mechanism and emerging links to disease.
Tran EJ, King MC, Corbett AH. Tran EJ, et al. Biochim Biophys Acta. 2014 Nov;1843(11):2784-2795. doi: 10.1016/j.bbamcr.2014.08.003. Epub 2014 Aug 9. Biochim Biophys Acta. 2014. PMID: 25116306 Free PMC article. Review.
References
- Al-Qattan MM, Shamseldin HE, Alkuraya FS. Familial dorsalization of the skin of the proximal palm and the instep of the sole of the foot. Gene. 2012;500:216–219. - PubMed
- Alcazar-Roman AR, Tran EJ, Guo S, Wente SR. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat. Cell Biol. 2006;8:711–716. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- T32 GM008320/GM/NIGMS NIH HHS/United States
- F31NS070431/NS/NINDS NIH HHS/United States
- T32CA119925/CA/NCI NIH HHS/United States
- 1DP2OD004483/OD/NIH HHS/United States
- R37GM051219/GM/NIGMS NIH HHS/United States
- F31 NS070431/NS/NINDS NIH HHS/United States
- DP2 OD004483/OD/NIH HHS/United States
- T32GM008320/GM/NIGMS NIH HHS/United States
- T32 CA119925/CA/NCI NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- R37 GM051219/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases