Reconstructing the population genetic history of the Caribbean - PubMed (original) (raw)
. 2013 Nov;9(11):e1003925.
doi: 10.1371/journal.pgen.1003925. Epub 2013 Nov 14.
Simon Gravel, Fouad Zakharia, Jacob L McCauley, Jake K Byrnes, Christopher R Gignoux, Patricia A Ortiz-Tello, Ricardo J Martínez, Dale J Hedges, Richard W Morris, Celeste Eng, Karla Sandoval, Suehelay Acevedo-Acevedo, Paul J Norman, Zulay Layrisse, Peter Parham, Juan Carlos Martínez-Cruzado, Esteban González Burchard, Michael L Cuccaro, Eden R Martin, Carlos D Bustamante
Affiliations
- PMID: 24244192
- PMCID: PMC3828151
- DOI: 10.1371/journal.pgen.1003925
Reconstructing the population genetic history of the Caribbean
Andrés Moreno-Estrada et al. PLoS Genet. 2013 Nov.
Abstract
The Caribbean basin is home to some of the most complex interactions in recent history among previously diverged human populations. Here, we investigate the population genetic history of this region by characterizing patterns of genome-wide variation among 330 individuals from three of the Greater Antilles (Cuba, Puerto Rico, Hispaniola), two mainland (Honduras, Colombia), and three Native South American (Yukpa, Bari, and Warao) populations. We combine these data with a unique database of genomic variation in over 3,000 individuals from diverse European, African, and Native American populations. We use local ancestry inference and tract length distributions to test different demographic scenarios for the pre- and post-colonial history of the region. We develop a novel ancestry-specific PCA (ASPCA) method to reconstruct the sub-continental origin of Native American, European, and African haplotypes from admixed genomes. We find that the most likely source of the indigenous ancestry in Caribbean islanders is a Native South American component shared among inland Amazonian tribes, Central America, and the Yucatan peninsula, suggesting extensive gene flow across the Caribbean in pre-Columbian times. We find evidence of two pulses of African migration. The first pulse--which today is reflected by shorter, older ancestry tracts--consists of a genetic component more similar to coastal West African regions involved in early stages of the trans-Atlantic slave trade. The second pulse--reflected by longer, younger tracts--is more similar to present-day West-Central African populations, supporting historical records of later transatlantic deportation. Surprisingly, we also identify a Latino-specific European component that has significantly diverged from its parental Iberian source populations, presumably as a result of small European founder population size. We demonstrate that the ancestral components in admixed genomes can be traced back to distinct sub-continental source populations with far greater resolution than previously thought, even when limited pre-Columbian Caribbean haplotypes have survived.
Conflict of interest statement
JKB is an employee of Ancestry.com. CDB is on the Scientific Advisory Board of Ancestry.com, 23andMe's “Roots into the Future” project, and Personalis, Inc. He is on the medical advisory board of Invitae and Med-tek. None of these entities played any role in the project or research results reported here.
Figures
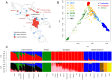
Figure 1. Population structure of Caribbean and neighboring populations.
A) Areas in red indicate countries of origin of newly genotyped admixed population samples and blue circles indicate new Venezuelan (underlined) and other previously published Native American samples. B) Principal Component Analysis and C) ADMIXTURE clustering analysis using the high-density dataset containing approximately 390 K autosomal SNP loci in common across admixed and reference panel populations. Unsupervised models assuming K = 3 and K = 8 ancestral clusters are shown. At K = 3, Caribbean admixed populations show extensive variation in continental ancestry proportions among and within groups. At K = 8, sub-continental components show differential proportions in recently admixed individuals. A Latino-specific European component accounts for the majority of the European ancestry among Caribbean Latinos and is exclusively shared with Iberian populations within Europe. Notably, this component is different from the two main gradients of ancestry differentiating southern from northern Europeans. Native Venezuelan components are present in higher proportions in admixed Colombians, Hondurans, and native Mayans.
Figure 2. Diagram of the analytical strategy used for reconstructing migration history and sub-continental ancestry in admixed genomes.
The starting point consists of genome-wide SNP data from family trios. Unrelated individuals are used to estimate global ancestry proportions with ADMIXTURE, whereas full trios are selected for BEAGLE phasing and PCA-based local ancestry estimation using continental reference samples. From here, two orthogonal analyses are performed: 1) Ancestry-specific regions of the genome are masked to separately apply PCA to European, African, and Native American haplotypes combined with large sub-continental reference panels of putative ancestral populations. We refer to this methodology as ancestry-specific PCA (ASPCA) and the code is packaged into the software PCAmask. 2) Continental-level local ancestry calls are used to estimate the tract length distribution per ancestry and population, which is then leveraged to test different demographic models of migration using Tracts software.
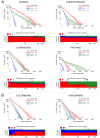
Figure 3. Demographic reconstruction since the onset of admixture in the Caribbean.
We used the length distribution of ancestry tracts within each population from A) insular and B) mainland Caribbean countries of origin. Scatter data points represent the observed distribution of ancestry tracts, and solid-colored lines represent the distribution from the model, with shaded areas indicating 68.3% confidence intervals. We used Markov models implemented in Tracts to test different demographic models for best fitting the observed data. Insular populations are best modeled when allowing for a second pulse of African ancestry, and mainland populations when a second pulse of European ancestry is allowed. Admixture time estimates (in number of generations ago), migration events, volume of migrants, and ancestry proportions over time are given for each population under the best-fitting model. The estimated age for the onset of admixture among insular populations is consistently older (i.e., 16–17) compared to that among mainland populations (i.e., 14).
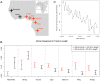
Figure 4. Sub-continental origin of Native American components in the Caribbean.
A) Ancestry-specific PCA analysis restricted to Native American segments from admixed Caribbean individuals (colored circles) and a reference panel of indigenous populations (gray symbols) from , grouped by sampling location. Darker symbols denote countries of origin with populations clustering closer to our Caribbean samples. Indigenous Colombian populations were classified into East and West of the Andes to ease the interpretation of their differential clustering in ASPCA. Population labels are shown for samples defining PC axes and representative clusters within locations. B) ADMIXTURE model for K = 16 ancestral clusters considering additional Latino samples, a representative subset of African and European source populations, and 52 Native American populations from , plus three additional Native Venezuelan tribes genotyped for this project. Vertical thin bars represent individuals and white spaces separate populations. Native American populations from are grouped according to linguistic families reported therein. Labels are shown for the populations representing the 12 Native American clusters identified at K = 16. Clusters involving multiple populations are identified by those with the highest membership values. C) Map showing the major indigenous components shared across the Caribbean basin as revealed by ADMIXTURE at K = 16 from B). Namely, Mesoamerican (blue), Chibchan (yellow), and South American (green). Colored bars represent individuals and their approximate sampling locations. Bars pooling genetically similar individuals from more than one population are plotted from left to right following north to south coordinates as listed by population labels. Guarani, Wichi, and Chane from north Argentina are pooled with Arara but only the location of the latter is shown to allow us to provide a zoomed view of the Caribbean region (see for the full map of sampling locations). The thick arrow represents schematically the most accepted origin of the Arawak expansion from South America into the Great Antilles around 2,500 years ago according to linguistic and archaeological evidence . Asterisks next to population labels denote Arawakan populations included in our reference panel. The thin arrow indicates gene flow between South America and Mesoamerica, possibly following a coastal or maritime route, accounting for the Mayan mixture and supporting pre-Columbian back migrations across the Caribbean.
Figure 5. Sub-continental origin of European haplotypes derived from admixed genomes.
ASPCA is applied to haploid genomes with >25% European ancestry derived from insular Caribbean (black symbols) and mainland populations (gray symbols) combined with a reference panel (colored labels) of 1,387 POPRES European samples with four grandparents from the same country , and 54 additional Iberian individuals (in yellow) from . PC1 values have been inverted and axes rotated 16 degrees counterclockwise to approximate the geographic orientation of population samples over Europe. Population codes are detailed in Table S1 and regions within Europe are labeled as in . Inset map: countries of origin for POPRES samples color-coded by region (areas not sampled in gray and Switzerland in intermediate shade of green to denote shared membership with EUR W, EUR C, and EUR S). Most Latino-derived European haplotypes cluster around the Iberian cluster. One of the two Haitian individuals included in the analysis clustered with French speaking Europeans (black arrow), in agreement with the colonial history of Haiti and illustrating the fine-scale resolution of our ASPCA approach.
Figure 6. Sub-continental origin of Afro-Caribbean haplotypes of different sizes.
A) Map of West Africa showing locations of reference panel populations. Samples in black are more likely to represent the origin of short ancestry tracts and those in red of long ancestry tracts, according to B) assignment probabilities for each putative ancestral population of being the source for short (<50 cM in black) and long (>50 cM in red) ancestry tracts. African ancestry tracts for Puerto Ricans are shown and results for all populations are available in Figure S16. C) Proportion of African ancestry of inferred Mandenka origin as a function of block size in the combined set of Caribbean genomes. By running PCAdmix within the previously inferred African segments, we obtained posterior probabilities for Mandenka versus Yoruba ancestry. Overall, we found evidence for a differential origin of the African lineages in present day Afro-Caribbean genomes, with shorter (and thus older) ancestry tracts tracing back to Far West Africa (represented by Mandenka and Brong), and longer tracts (and thus younger) tracing back to Central West Africa.
Similar articles
- Genomic Insights into the Ancestry and Demographic History of South America.
Homburger JR, Moreno-Estrada A, Gignoux CR, Nelson D, Sanchez E, Ortiz-Tello P, Pons-Estel BA, Acevedo-Vasquez E, Miranda P, Langefeld CD, Gravel S, Alarcón-Riquelme ME, Bustamante CD. Homburger JR, et al. PLoS Genet. 2015 Dec 4;11(12):e1005602. doi: 10.1371/journal.pgen.1005602. eCollection 2015 Dec. PLoS Genet. 2015. PMID: 26636962 Free PMC article. - Reconstructing Native American migrations from whole-genome and whole-exome data.
Gravel S, Zakharia F, Moreno-Estrada A, Byrnes JK, Muzzio M, Rodriguez-Flores JL, Kenny EE, Gignoux CR, Maples BK, Guiblet W, Dutil J, Via M, Sandoval K, Bedoya G; 1000 Genomes Project; Oleksyk TK, Ruiz-Linares A, Burchard EG, Martinez-Cruzado JC, Bustamante CD. Gravel S, et al. PLoS Genet. 2013;9(12):e1004023. doi: 10.1371/journal.pgen.1004023. Epub 2013 Dec 26. PLoS Genet. 2013. PMID: 24385924 Free PMC article. - Genome-wide Ancestry and Demographic History of African-Descendant Maroon Communities from French Guiana and Suriname.
Fortes-Lima C, Gessain A, Ruiz-Linares A, Bortolini MC, Migot-Nabias F, Bellis G, Moreno-Mayar JV, Restrepo BN, Rojas W, Avendaño-Tamayo E, Bedoya G, Orlando L, Salas A, Helgason A, Gilbert MTP, Sikora M, Schroeder H, Dugoujon JM. Fortes-Lima C, et al. Am J Hum Genet. 2017 Nov 2;101(5):725-736. doi: 10.1016/j.ajhg.2017.09.021. Am J Hum Genet. 2017. PMID: 29100086 Free PMC article. - Anthropological genetic insights on Caribbean population history.
Nieves-Colón MA. Nieves-Colón MA. Evol Anthropol. 2022 May;31(3):118-137. doi: 10.1002/evan.21935. Epub 2022 Jan 21. Evol Anthropol. 2022. PMID: 35060661 Review. - The Genetic Diversity of the Americas.
Adhikari K, Chacón-Duque JC, Mendoza-Revilla J, Fuentes-Guajardo M, Ruiz-Linares A. Adhikari K, et al. Annu Rev Genomics Hum Genet. 2017 Aug 31;18:277-296. doi: 10.1146/annurev-genom-083115-022331. Annu Rev Genomics Hum Genet. 2017. PMID: 28859572 Review.
Cited by
- Nested admixture during and after the Trans-Atlantic Slave Trade on the island of São Tomé.
Ciccarella M, Laurent R, Szpiech ZA, Patin E, Dessarps-Freichey F, Utgé J, Lémée L, Semo A, Rocha J, Verdu P. Ciccarella M, et al. bioRxiv [Preprint]. 2024 Oct 23:2024.10.21.619344. doi: 10.1101/2024.10.21.619344. bioRxiv. 2024. PMID: 39484499 Free PMC article. Preprint. - Differential prediction performance between Caribbean- and Mainland-subgroups using state-of-the-art polygenic risk scores for coronary heart disease: Findings from the Hispanic Community Health Study/Study of Latinos (HCHS/SOL).
Hutten CG, Boehm FJ, Smith JA, Spitzer BW, Wassertheil-Smoller S, Isasi CR, Cai J, Unkart JT, Sun J, Persky V, Daviglus ML, Sofer T, Argos M. Hutten CG, et al. medRxiv [Preprint]. 2024 Sep 27:2024.09.25.24313663. doi: 10.1101/2024.09.25.24313663. medRxiv. 2024. PMID: 39399039 Free PMC article. Preprint. - Gene profiling of Epstein-Barr Virus and human endogenous retrovirus in peripheral blood mononuclear cells of SLE patients: immune response implications.
Lemus YB, Martínez GA, Lugo LP, Escorcia LG, Peñata EZ, Llanos NS, Bonfanti AC, Acosta-Hoyos AJ, Quiroz EN. Lemus YB, et al. Sci Rep. 2024 Aug 30;14(1):20236. doi: 10.1038/s41598-024-70913-6. Sci Rep. 2024. PMID: 39215087 Free PMC article. - Global and Local Ancestry and its Importance: A Review.
Goli RC, Chishi KG, Ganguly I, Singh S, Dixit SP, Rathi P, Diwakar V, Sree C C, Limbalkar OM, Sukhija N, Kanaka KK. Goli RC, et al. Curr Genomics. 2024;25(4):237-260. doi: 10.2174/0113892029298909240426094055. Epub 2024 May 9. Curr Genomics. 2024. PMID: 39156729 Free PMC article. Review. - Social determinants of health but not global genetic ancestry predict dementia prevalence in Latin America.
Llibre-Guerra JJ, Jiang M, Acosta I, Sosa AL, Acosta D, Jimenez-Velasquez IZ, Guerra M, Salas A, Rodriguez Salgado AM, Llibre-Guerra JC, Sánchez ND, Prina M, Renton A, Albanese E, Yokoyama JS, Llibre Rodriguez JJ. Llibre-Guerra JJ, et al. Alzheimers Dement. 2024 Jul;20(7):4828-4840. doi: 10.1002/alz.14041. Epub 2024 Jun 5. Alzheimers Dement. 2024. PMID: 38837526 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM090087/GM/NIGMS NIH HHS/United States
- P60MD006902/MD/NIMHD NIH HHS/United States
- P60 MD006902/MD/NIMHD NIH HHS/United States
- 1R01GM090087/GM/NIGMS NIH HHS/United States
- T32 GM007175/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources