Regulation of NO synthesis, local inflammation, and innate immunity to pathogens by BET family proteins - PubMed (original) (raw)
Regulation of NO synthesis, local inflammation, and innate immunity to pathogens by BET family proteins
Sebastian Wienerroither et al. Mol Cell Biol. 2014 Feb.
Abstract
Transcriptional activation of the Nos2 gene, encoding inducible nitric oxide synthase (iNOS), during infection or inflammation requires coordinate assembly of an initiation complex by the transcription factors NF-κB and type I interferon-activated ISGF3. Here we show that infection of macrophages with the intracellular bacterial pathogen Listeria monocytogenes caused binding of the BET proteins Brd2, Brd3, and, most prominently, Brd4 to the Nos2 promoter and that a profound reduction of Nos2 expression occurred in the presence of the BET inhibitor JQ1. RNA polymerase activity at the Nos2 gene was regulated through Brd-mediated C-terminal domain (CTD) phosphorylation at serine 5. Underscoring the critical importance of Brd for the regulation of immune responses, application of JQ1 reduced NO production in mice infected with L. monocytogenes, as well as innate resistance to L. monocytogenes and influenza virus. In a murine model of inflammatory disease, JQ1 treatment increased the colitogenic activity of dextran sodium sulfate (DSS). The data presented in our study suggest that BET protein inhibition in a clinical setting poses the risk of altering the innate immune response to infectious or inflammatory challenge.
Figures
FIG 1
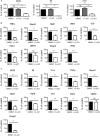
Sensitivity of Listeria monocytogenes-induced gene expression to BET protein inhibition with JQ1. Bone marrow-derived macrophages (BMDM) were infected with L. monocytogenes for 4 h (A and B) or treated with a combination of heat-killed L. monocytogenes and IFN-β (C). Where indicated, 250 nM JQ1 was added 1 h before infection and left in the culture medium during infection. Gene expression was determined by Q-PCR. Values represent means and standard errors for three independent biological replicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
FIG 2
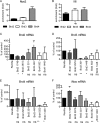
Recruitment of BET proteins to the Nos2 promoter and inhibition of Nos2 expression by Brd shRNAs. All experiments were carried out with BMDM. (A and B) The cells were treated with a combination of heat-killed Listeria and IFN-β, followed by ChIP with the indicated antibodies and amplification of the Nos2 promoter region (A) or IL-6 promoter region (B), including the TSS, by Q-PCR. n = 5 (A) or 3 (B). (C to E) BMDM isolated from Rosa26-rtTA-M2 transgenic mice (49) were spin infected as described in Materials and Methods with a retrovirus expressing tet-inducible Brd shRNA. shRNA expression was induced 2 days after infection by adding 1 μg/ml dox to the medium, and shRNA-expressing (Turbo-GFP+) cells were FACS sorted after 5 days of dox treatment. The efficacy of the Brd knockdown in cells expressing shRNA was determined by Q-PCR (n = 3). (F) BMDM obtained as described for panels C to E were analyzed for shRNA-mediated inhibition of Nos2 expression by Q-PCR (n = 3). *, P < 0.05; **, P < 0.01; ns, not significant.
FIG 3
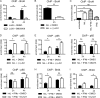
Impact of BET, IKKβ, or HDAC inhibition on the recruitment of Brd4 and NF-κB p65 to Nos2 chromatin. (A and B) BMDM were infected with Listeria monocytogenes strain Lo28 for the indicated time in the presence or absence of the IKKβ inhibitor BI605906 at 3 μM (A) or 250 nM JQ1 (B), followed by ChIP with antibodies to Brd4. (C) BMDM were treated with heat-killed L. monocytogenes (hkL), IFN-β, or a combination of both, and Brd4 binding to the Nos2 promoter was measured as described for panel A. (D and E) The cells were treated with either heat-killed L. monocytogenes (D) or a combination of heat-killed Listeria and IFN-β (E) in the presence or absence of 250 nM JQ1, followed by ChIP with antibodies to NF-κB p65 and amplification of the Nos2 promoter region, including the TSS, by Q-PCR. (F and G) The cells were treated with a combination of heat-killed Listeria and IFN-β in the presence or absence of the histone deacetylase inhibitor MS-275 at 2 μM (F) or Ex-527 at 10 μM (G), followed by ChIP with antibodies to NF-κB p65 and amplification of the Nos2 promoter region, including the TSS, by Q-PCR. (H and I) Treatment was the same as in panels F and G, but ChIP was done with antibodies to Brd4. The Nos2 promoter region, including the TSS, was amplified by Q-PCR. n ≥ 3 for all experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
FIG 4
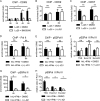
Impact of BET inhibition on CDK7, CDK9, and Pol II association with the Nos2 promoter and on phosphorylation of the Pol II CTD. (A) Recruitment of CDK9 to the Nos2 promoter of L. monocytogenes (Lo28)-infected BMDM as determined by ChIP and Q-PCR amplification of the proximal Nos2 promoter. White bars indicate CDK9 recruitment in the presence of the IKKβ inhibitor BI605906. (B and C) Impact of BET inhibition by JQ1 on the recruitment of CDK9 (B) and CDK7 (C). Untreated and L. monocytogenes-infected BMDM were subjected to ChIP with antibodies to CDK9 and CDK7. Where indicated, BET proteins were additionally inhibited by treatment with 250 nM JQ1. (D, E, and G) Impact of BET inhibition on recruitment of Pol II (D) and S2-phosphorylated (E) or S5-phosphorylated (G) Pol II to the Nos2 promoter or exonic regions. BMDM were left untreated or treated with a combination of heat-killed L. monocytogenes and IFN-β (black bars). Where indicated, BET proteins were additionally inhibited by treatment with 250 nM JQ1 (white bars). S2- or S5-phosphorylated Pol II association was determined by ChIP. (F) Ratio of S2-phosphorylated Pol II and total Pol II at different regions of the Nos2 gene. (H) Ratio of S5-phosphorylated Pol II and total Pol II at different regions of the Nos2 gene. Values represent means and standard errors for biological replicates. n = 3 (B, F, and H) or 4 (A, C, D, E, and G). *, P < 0.05; **, P < 0.01; ns, not significant.
FIG 5
Impact of Brd4 inhibition on NO production and innate immunity to Listeria monocytogenes. (A) Untreated or JQ1-treated mice (daily injections of 50 mg/kg i.p.) were infected intraperitoneally with L. monocytogenes (Lo28). Twenty-four hours after infection, the spleen was removed. Splenic leukocytes were cultured for 36 h, and supernatants were collected for the determination of NO with Griess reagent (n = 5 per group). (B) BMDM were left untreated or treated with 250 nM JQ1. The cells were infected with L. monocytogenes for the indicated times, followed by an assessment of intracellular L. monocytogenes by CFU assay. The experiment is representative of more than three independent biological replicates. (C to G) Untreated mice (n = 5) or mice treated with JQ1 as in panel A (n = 5) were infected intraperitoneally with L. monocytogenes. Infected mice were analyzed after 48 h for the bacterial burdens in the spleen and liver (C and D) or for survival over a 10-day observation period (E to G) (n = 10 per group; data from three independent experiments were combined). Panels F and G show data for animals additionally treated intraperitoneally with 0.5 or 1 μg (n = 10 per group), respectively, of TNF-α before infection with L. monocytogenes to test the cytokine's ability to rescue the JQ1 effect. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
FIG 6
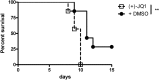
Effect of BET inhibition on resistance to influenza virus. Untreated or JQ1-treated mice (daily injections at 50 mg/kg) were infected with 500 PFU of a mouse-adapted influenza A virus (H1N1 subtype; strain WSN/33), and survival was monitored over 15 days (n = 8; data from two independent experiments with n = 4 were combined). **, P < 0.01.
FIG 7
Effect of BET inhibition on DSS-induced colitis. (A to D) Untreated or JQ1-treated mice (daily injections of 50 mg/kg i.p.) were given 2% DSS in their drinking water or kept on regular drinking water over a 7-day period. Colitis was assessed by weight loss over 10 days (A) or 7 days (B) (see the text for further information), shortening of the colon (C), and pathology score (D) (n = 8; data from two independent experiments with n = 4 were combined). (E and F) Histological examination of the colon mucosa on day 7 of the DSS treatment protocol in the absence (E) or presence (F) of JQ1. Micrographs represent thin sections of paraffin-embedded tissue stained with hematoxylin and eosin. (G) FITC-labeled dextran (molecular mass of 3,000 to 5,000 Da) was given to mice via gavage. The appearance of fluorescent material in the blood was measured 3 h later. (H to L) Expression of the indicated genes was measured by Q-PCR following mRNA extraction from the colon mucosa. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Similar articles
- BET protein function is required for inflammation: Brd2 genetic disruption and BET inhibitor JQ1 impair mouse macrophage inflammatory responses.
Belkina AC, Nikolajczyk BS, Denis GV. Belkina AC, et al. J Immunol. 2013 Apr 1;190(7):3670-8. doi: 10.4049/jimmunol.1202838. Epub 2013 Feb 18. J Immunol. 2013. PMID: 23420887 Free PMC article. - Nitric oxide and KLF4 protein epigenetically modify class II transactivator to repress major histocompatibility complex II expression during Mycobacterium bovis bacillus Calmette-Guerin infection.
Ghorpade DS, Holla S, Sinha AY, Alagesan SK, Balaji KN. Ghorpade DS, et al. J Biol Chem. 2013 Jul 12;288(28):20592-606. doi: 10.1074/jbc.M113.472183. Epub 2013 Jun 3. J Biol Chem. 2013. PMID: 23733190 Free PMC article. - Nonconventional initiation complex assembly by STAT and NF-kappaB transcription factors regulates nitric oxide synthase expression.
Farlik M, Reutterer B, Schindler C, Greten F, Vogl C, Müller M, Decker T. Farlik M, et al. Immunity. 2010 Jul 23;33(1):25-34. doi: 10.1016/j.immuni.2010.07.001. Immunity. 2010. PMID: 20637660 Free PMC article. - Nitric oxide synthase in innate and adaptive immunity: an update.
Bogdan C. Bogdan C. Trends Immunol. 2015 Mar;36(3):161-78. doi: 10.1016/j.it.2015.01.003. Epub 2015 Feb 13. Trends Immunol. 2015. PMID: 25687683 Review. - WHAT do viruses BET on?
Weidner-Glunde M, Ottinger M, Schulz TF. Weidner-Glunde M, et al. Front Biosci (Landmark Ed). 2010 Jan 1;15(2):537-49. doi: 10.2741/3632. Front Biosci (Landmark Ed). 2010. PMID: 20036832 Review.
Cited by
- Epigenetic drug discovery: breaking through the immune barrier.
Tough DF, Tak PP, Tarakhovsky A, Prinjha RK. Tough DF, et al. Nat Rev Drug Discov. 2016 Dec;15(12):835-853. doi: 10.1038/nrd.2016.185. Epub 2016 Oct 21. Nat Rev Drug Discov. 2016. PMID: 27765940 Review. - Selective BET-bromodomain inhibition by JQ1 suppresses dendritic cell maturation and antigen-specific T-cell responses.
Remke N, Bisht S, Oberbeck S, Nolting J, Brossart P. Remke N, et al. Cancer Immunol Immunother. 2021 Jan;70(1):107-121. doi: 10.1007/s00262-020-02665-x. Epub 2020 Jul 10. Cancer Immunol Immunother. 2021. PMID: 32651619 Free PMC article. - BRD4 Regulates Glycolysis-Dependent Nos2 Expression in Macrophages Upon H pylori Infection.
Modi N, Chen Y, Dong X, Hu X, Lau GW, Wilson KT, Peek RM Jr, Chen LF. Modi N, et al. Cell Mol Gastroenterol Hepatol. 2024;17(2):292-308.e1. doi: 10.1016/j.jcmgh.2023.10.001. Epub 2023 Oct 10. Cell Mol Gastroenterol Hepatol. 2024. PMID: 37820788 Free PMC article. - BET N-terminal bromodomain inhibition selectively blocks Th17 cell differentiation and ameliorates colitis in mice.
Cheung K, Lu G, Sharma R, Vincek A, Zhang R, Plotnikov AN, Zhang F, Zhang Q, Ju Y, Hu Y, Zhao L, Han X, Meslamani J, Xu F, Jaganathan A, Shen T, Zhu H, Rusinova E, Zeng L, Zhou J, Yang J, Peng L, Ohlmeyer M, Walsh MJ, Zhang DY, Xiong H, Zhou MM. Cheung K, et al. Proc Natl Acad Sci U S A. 2017 Mar 14;114(11):2952-2957. doi: 10.1073/pnas.1615601114. Epub 2017 Mar 6. Proc Natl Acad Sci U S A. 2017. PMID: 28265070 Free PMC article. - BET proteins are a key component of immunoglobulin gene expression.
Shim JM, Lee JS, Russell KE, Wiegman CH, Barnes PJ, Fear D, Adcock IM, Durham AL. Shim JM, et al. Epigenomics. 2017 Apr;9(4):393-406. doi: 10.2217/epi-2016-0147. Epub 2017 Mar 21. Epigenomics. 2017. PMID: 28322577 Free PMC article.
References
- Seki E, Tsutsui H, Tsuji NM, Hayashi N, Adachi K, Nakano H, Futatsugi-Yumikura S, Takeuchi O, Hoshino K, Akira S, Fujimoto J, Nakanishi K. 2002. Critical roles of myeloid differentiation factor 88-dependent proinflammatory cytokine release in early phase clearance of Listeria monocytogenes in mice. J. Immunol. 169:3863–3868 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous