The basic building blocks and evolution of CRISPR-CAS systems - PubMed (original) (raw)
Review
The basic building blocks and evolution of CRISPR-CAS systems
Kira S Makarova et al. Biochem Soc Trans. 2013 Dec.
Abstract
CRISPR (clustered regularly interspaced short palindromic repeats)-Cas (CRISPR-associated) is an adaptive immunity system in bacteria and archaea that functions via a distinct self/non-self recognition mechanism that involves unique spacers homologous with viral or plasmid DNA and integrated into the CRISPR loci. Most of the Cas proteins evolve under relaxed purifying selection and some underwent dramatic structural rearrangements during evolution. In many cases, CRISPR-Cas system components are replaced either by homologous or by analogous proteins or domains in some bacterial and archaeal lineages. However, recent advances in comparative sequence analysis, structural studies and experimental data suggest that, despite this remarkable evolutionary plasticity, all CRISPR-Cas systems employ the same architectural and functional principles, and given the conservation of the principal building blocks, share a common ancestry. We review recent advances in the understanding of the evolution and organization of CRISPR-Cas systems. Among other developments, we describe for the first time a group of archaeal cas1 gene homologues that are not associated with CRISPR-Cas loci and are predicted to be involved in functions other than adaptive immunity.
Figures
Figure 1. The principal building blocks of CRISPR–Cas system types
Gene names and other identifiers follow the current nomenclature and classification [7,30]. An asterisk indicates the putative small subunit that might be fused to the large subunit in several Type I subtypes [30]. Dispensable genes are indicated by broken outlines.
Figure 2. The Cas1 family evolution and functional associations
(A) The Cas1 family phylogeny. The PSIBLAST program [50] was used to retrieve homologues of the Cas1 family from 2262 completely sequenced genomes in the NCBI nr database. The BLASTCLUST program [51] (length coverage cut-off, 0.8; score density threshold, 1.0) was used to select 205 representative sequences. The multiple alignment was built using the MUSCLE program [52] with default parameters. The FastTree program [53] (JTT evolutionary model, discrete gamma model for site rates with 20 rate categories) was used for tree reconstruction. The branches are coloured according to the assignment of Cas1 genes to system subtypes based on the analysis of ten upstream and ten downstream genes. X denotes systems of unknown type or those that are predicted to be derivatives of the respective system (when coloured). (B) Phylogeny of the two solo Cas1 families. The conserved gene neighbourhood of the second group is shown underneath the trees. (C) Cas1 fusions and operonic associations.
Figure 3. Putative common architectural organization of central Cascade components and the evolutionary scenario for the Cascade origin
(A) The subdomain architectures of Cse1, Cas10 and Cas7. The similarly coloured domains could be homologous but dramatically rearranged. The small subunit of subtype III-B that could be homologous with C-terminal helical domain of Cas10 is also shown. (B) Evolutionary scenario for the origin of the Cascade-like complex. Homologous domains or subdomains are colour-coded and identified by a family name, which follow the modified classification [30]. This scenario is a modification of the previously described one [28].
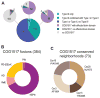
Figure 4. COG1517, a dormancy/programmed cell death system component associated with CRISPR–Cas systems of Type III
(A) CRISPR–Cas-associated ‘effector’ domains. The bottom pie chart shows the breakdown of 314 CRISPR–Cas-positive genomes by the system types. The pie charts at the top show the breakdown of the respective sectors with respect to the presence of at least one COG1517 family protein and the presence or absence of an ‘effector/toxin’ domain in these proteins. (B) Effector/toxin domain fusions. HD and PD-(D/E)xK are DNA nucleases, and PIN, RelE and HEPN are ribonucleases. The number of fusions identified is indicated in parentheses. (C) Uncharacterized gene families associated with COG1517 genes in putative operons. The number of operons predicted is indicated in parentheses.
Similar articles
- CRISPR-Cas: evolution of an RNA-based adaptive immunity system in prokaryotes.
Koonin EV, Makarova KS. Koonin EV, et al. RNA Biol. 2013 May;10(5):679-86. doi: 10.4161/rna.24022. Epub 2013 Feb 25. RNA Biol. 2013. PMID: 23439366 Free PMC article. Review. - Viral diversity threshold for adaptive immunity in prokaryotes.
Weinberger AD, Wolf YI, Lobkovsky AE, Gilmore MS, Koonin EV. Weinberger AD, et al. mBio. 2012 Dec 4;3(6):e00456-12. doi: 10.1128/mBio.00456-12. mBio. 2012. PMID: 23221803 Free PMC article. - Adaptation in CRISPR-Cas Systems.
Sternberg SH, Richter H, Charpentier E, Qimron U. Sternberg SH, et al. Mol Cell. 2016 Mar 17;61(6):797-808. doi: 10.1016/j.molcel.2016.01.030. Epub 2016 Mar 3. Mol Cell. 2016. PMID: 26949040 Review. - CRISPR-Cas systems: beyond adaptive immunity.
Westra ER, Buckling A, Fineran PC. Westra ER, et al. Nat Rev Microbiol. 2014 May;12(5):317-26. doi: 10.1038/nrmicro3241. Epub 2014 Apr 7. Nat Rev Microbiol. 2014. PMID: 24704746 Review. - How type II CRISPR-Cas establish immunity through Cas1-Cas2-mediated spacer integration.
Xiao Y, Ng S, Nam KH, Ke A. Xiao Y, et al. Nature. 2017 Oct 5;550(7674):137-141. doi: 10.1038/nature24020. Epub 2017 Sep 4. Nature. 2017. PMID: 28869593 Free PMC article.
Cited by
- CRISPR-Cas9: A History of Its Discovery and Ethical Considerations of Its Use in Genome Editing.
Gostimskaya I. Gostimskaya I. Biochemistry (Mosc). 2022 Aug;87(8):777-788. doi: 10.1134/S0006297922080090. Biochemistry (Mosc). 2022. PMID: 36171658 Free PMC article. Review. - Annotation and Classification of CRISPR-Cas Systems.
Makarova KS, Koonin EV. Makarova KS, et al. Methods Mol Biol. 2015;1311:47-75. doi: 10.1007/978-1-4939-2687-9_4. Methods Mol Biol. 2015. PMID: 25981466 Free PMC article. Review. - Type I CRISPR-Cas-mediated microbial gene editing and regulation.
Xu Z, Chen S, Wu W, Wen Y, Cao H. Xu Z, et al. AIMS Microbiol. 2023 Dec 18;9(4):780-800. doi: 10.3934/microbiol.2023040. eCollection 2023. AIMS Microbiol. 2023. PMID: 38173969 Free PMC article. Review. - Visualization and prediction of CRISPR incidence in microbial trait-space to identify drivers of antiviral immune strategy.
Weissman JL, Laljani RMR, Fagan WF, Johnson PLF. Weissman JL, et al. ISME J. 2019 Oct;13(10):2589-2602. doi: 10.1038/s41396-019-0411-2. Epub 2019 Jun 25. ISME J. 2019. PMID: 31239539 Free PMC article. - Evolution of RNA- and DNA-guided antivirus defense systems in prokaryotes and eukaryotes: common ancestry vs convergence.
Koonin EV. Koonin EV. Biol Direct. 2017 Feb 10;12(1):5. doi: 10.1186/s13062-017-0177-2. Biol Direct. 2017. PMID: 28187792 Free PMC article. Review.
References
- Barrangou R, Horvath P. CRISPR: new horizons in phage resistance and strain identification. Annu Rev Food Sci Technol. 2012;3:143–162. - PubMed
- Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources