The RNA- and DNA-targeting CRISPR-Cas immune systems of Pyrococcus furiosus - PubMed (original) (raw)
Review
The RNA- and DNA-targeting CRISPR-Cas immune systems of Pyrococcus furiosus
Rebecca M Terns et al. Biochem Soc Trans. 2013 Dec.
Abstract
Using the hyperthermophile Pyrococcus furiosus, we have delineated several key steps in CRISPR (clustered regularly interspaced short palindromic repeats)-Cas (CRISPR-associated) invader defence pathways. P. furiosus has seven transcriptionally active CRISPR loci that together encode a total of 200 crRNAs (CRISPR RNAs). The 27 Cas proteins in this organism represent three distinct pathways and are primarily encoded in two large gene clusters. The Cas6 protein dices CRISPR locus transcripts to generate individual invader-targeting crRNAs. The mature crRNAs include a signature sequence element (the 5' tag) derived from the CRISPR locus repeat sequence that is important for function. crRNAs are tailored into distinct species and integrated into three distinct crRNA-Cas protein complexes that are all candidate effector complexes. The complex formed by the Cmr [Cas module RAMP (repeat-associated mysterious proteins)] (subtype III-B) proteins cleaves complementary target RNAs and can be programmed to cleave novel target RNAs in a prokaryotic RNAi-like manner. Evidence suggests that the other two CRISPR-Cas systems in P. furiosus, Csa (Cas subtype Apern) (subtype I-A) and Cst (Cas subtype Tneap) (subtype I-B), target invaders at the DNA level. Studies of the CRISPR-Cas systems from P. furiosus are yielding fundamental knowledge of mechanisms of crRNA biogenesis and silencing for three of the diverse CRISPR-Cas pathways, and reveal that organisms such as P. furiosus possess an arsenal of multiple RNA-guided mechanisms to resist diverse invaders. Our knowledge of the fascinating CRISPR-Cas pathways is leading in turn to our ability to co-opt these systems for exciting new biomedical and biotechnological applications.
Figures
Figure 1. P. furiosus CRISPR–Cas systems
(A) Distribution of the CRISPR loci (Cr1, Cr2, Cr4–Cr8; red) and two major cas gene clusters (Cas; blue) throughout the P. furiosus genome. (B) CRISPRs comprise leader regions (L) preceding series of repeat (black) and spacer (coloured) elements. The actual numbers of spacers are indicated. The consensus crRNA repeat sequence found in the seven CRISPR loci is shown above with the Cas6-binding and cleavage sites, and the 8-nt 5′ tag crRNA sequence indicated. (C) Genome organization of predicted P. furiosus (Pf) cas genes. Subtype III-B Cmr (blue), subtype I-B Cst (yellow), subtype I-A Csa (green) and core cas genes (grey) are indicated, and recent cas gene superfamily designations [15] are indicated below [14]. The known or predicted general function of the core cas genes is indicated.
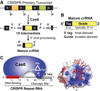
Figure 2. Cas6 and crRNA biogenesis
(A) Cas6 cleaves within the repeat regions of the CRISPR transcript to liberate the individual embedded crRNAs. The products of Cas6 cleavage (1× crRNAs) undergo 3′ end trimming. (B) The architecture of a mature crRNA. This abundant crRNA form includes the 5′ repeat-derived tag and complete guide sequence. Other mature crRNAs in P. furiosus contain some repeat sequences at their 3′ ends or slightly less than full-length guide sequences (not depicted). (C) Cas6 is an endoribonuclease critical for crRNA biogenesis. Cas6 interacts with the 5′ region of the CRISPR repeat RNA (red box) and cleaves at a distal site using an active site composed of three key amino acids (indicated). The Cas6-binding region in the crRNA interacts with the positively charged cleft of Cas6.
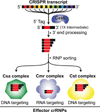
Figure 3. crRNAs are loaded into three distinct crRNP immune effector complexes in P. furiosus
Following release by Cas6 and further 3′ end trimming, crRNAs of specific size formed from all seven CRISPR loci become incorporated into the three distinct crRNP complexes containing Cmr, Csa or Cst Cas proteins. There is evidence that Cmr crRNPs cleave target RNAs, whereas Csa and Cst complexes cleave invader DNA.
Similar articles
- Three CRISPR-Cas immune effector complexes coexist in Pyrococcus furiosus.
Majumdar S, Zhao P, Pfister NT, Compton M, Olson S, Glover CV 3rd, Wells L, Graveley BR, Terns RM, Terns MP. Majumdar S, et al. RNA. 2015 Jun;21(6):1147-58. doi: 10.1261/rna.049130.114. Epub 2015 Apr 22. RNA. 2015. PMID: 25904135 Free PMC article. - DNA targeting by the type I-G and type I-A CRISPR-Cas systems of Pyrococcus furiosus.
Elmore J, Deighan T, Westpheling J, Terns RM, Terns MP. Elmore J, et al. Nucleic Acids Res. 2015 Dec 2;43(21):10353-63. doi: 10.1093/nar/gkv1140. Epub 2015 Oct 30. Nucleic Acids Res. 2015. PMID: 26519471 Free PMC article. - Hot and crispy: CRISPR-Cas systems in the hyperthermophile Sulfolobus solfataricus.
Zhang J, White MF. Zhang J, et al. Biochem Soc Trans. 2013 Dec;41(6):1422-6. doi: 10.1042/BST20130031. Biochem Soc Trans. 2013. PMID: 24256231 Review. - Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs.
Hale CR, Majumdar S, Elmore J, Pfister N, Compton M, Olson S, Resch AM, Glover CV 3rd, Graveley BR, Terns RM, Terns MP. Hale CR, et al. Mol Cell. 2012 Feb 10;45(3):292-302. doi: 10.1016/j.molcel.2011.10.023. Epub 2012 Jan 5. Mol Cell. 2012. PMID: 22227116 Free PMC article. - Approaches to study CRISPR RNA biogenesis and the key players involved.
Behler J, Hess WR. Behler J, et al. Methods. 2020 Feb 1;172:12-26. doi: 10.1016/j.ymeth.2019.07.015. Epub 2019 Jul 17. Methods. 2020. PMID: 31325492 Review.
Cited by
- _O_6-alkylguanine-DNA Alkyltransferases in Microbes Living on the Edge: From Stability to Applicability.
Mattossovich R, Merlo R, Miggiano R, Valenti A, Perugino G. Mattossovich R, et al. Int J Mol Sci. 2020 Apr 20;21(8):2878. doi: 10.3390/ijms21082878. Int J Mol Sci. 2020. PMID: 32326075 Free PMC article. Review. - Role of free DNA ends and protospacer adjacent motifs for CRISPR DNA uptake in Pyrococcus furiosus.
Shiimori M, Garrett SC, Chambers DP, Glover CVC 3rd, Graveley BR, Terns MP. Shiimori M, et al. Nucleic Acids Res. 2017 Nov 2;45(19):11281-11294. doi: 10.1093/nar/gkx839. Nucleic Acids Res. 2017. PMID: 29036456 Free PMC article. - A journey down to hell: new thermostable protein-tags for biotechnology at high temperatures.
Mattossovich R, Merlo R, Fontana A, d'Ippolito G, Terns MP, Watts EA, Valenti A, Perugino G. Mattossovich R, et al. Extremophiles. 2020 Jan;24(1):81-91. doi: 10.1007/s00792-019-01134-3. Epub 2019 Sep 25. Extremophiles. 2020. PMID: 31555904 Free PMC article. - The nuts and bolts of the Haloferax CRISPR-Cas system I-B.
Maier LK, Stachler AE, Brendel J, Stoll B, Fischer S, Haas KA, Schwarz TS, Alkhnbashi OS, Sharma K, Urlaub H, Backofen R, Gophna U, Marchfelder A. Maier LK, et al. RNA Biol. 2019 Apr;16(4):469-480. doi: 10.1080/15476286.2018.1460994. Epub 2018 May 21. RNA Biol. 2019. PMID: 29649958 Free PMC article. - Three CRISPR-Cas immune effector complexes coexist in Pyrococcus furiosus.
Majumdar S, Zhao P, Pfister NT, Compton M, Olson S, Glover CV 3rd, Wells L, Graveley BR, Terns RM, Terns MP. Majumdar S, et al. RNA. 2015 Jun;21(6):1147-58. doi: 10.1261/rna.049130.114. Epub 2015 Apr 22. RNA. 2015. PMID: 25904135 Free PMC article.
References
- Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 2013;82:237–266. - PubMed
- Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. - PubMed
- Westra ER, Swarts DC, Staals RH, Jore MM, Brouns SJ, van der Oost J. The CRISPRs, they are a-changin’: how prokaryotes generate adaptive immunity. Annu. Rev. Genet. 2012;46:311–339. - PubMed
- Bhaya D, Davison M, Barrangou R. CRISPR–Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 2011;45:273–297. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources