A modular organization of the human intestinal mucosal microbiota and its association with inflammatory bowel disease - PubMed (original) (raw)
. 2013 Nov 19;8(11):e80702.
doi: 10.1371/journal.pone.0080702. eCollection 2013.
Xiaoxiao Li, Laura Wegener Parfrey, Bennett Roth, Andrew Ippoliti, Bo Wei, James Borneman, Dermot P B McGovern, Daniel N Frank, Ellen Li, Steve Horvath, Rob Knight, Jonathan Braun
Affiliations
- PMID: 24260458
- PMCID: PMC3834335
- DOI: 10.1371/journal.pone.0080702
A modular organization of the human intestinal mucosal microbiota and its association with inflammatory bowel disease
Maomeng Tong et al. PLoS One. 2013.
Abstract
Abnormalities of the intestinal microbiota are implicated in the pathogenesis of Crohn's disease (CD) and ulcerative colitis (UC), two spectra of inflammatory bowel disease (IBD). However, the high complexity and low inter-individual overlap of intestinal microbial composition are formidable barriers to identifying microbial taxa representing this dysbiosis. These difficulties might be overcome by an ecologic analytic strategy to identify modules of interacting bacteria (rather than individual bacteria) as quantitative reproducible features of microbial composition in normal and IBD mucosa. We sequenced 16S ribosomal RNA genes from 179 endoscopic lavage samples from different intestinal regions in 64 subjects (32 controls, 16 CD and 16 UC patients in clinical remission). CD and UC patients showed a reduction in phylogenetic diversity and shifts in microbial composition, comparable to previous studies using conventional mucosal biopsies. Analysis of weighted co-occurrence network revealed 5 microbial modules. These modules were unprecedented, as they were detectable in all individuals, and their composition and abundance was recapitulated in an independent, biopsy-based mucosal dataset 2 modules were associated with healthy, CD, or UC disease states. Imputed metagenome analysis indicated that these modules displayed distinct metabolic functionality, specifically the enrichment of oxidative response and glycan metabolism pathways relevant to host-pathogen interaction in the disease-associated modules. The highly preserved microbial modules accurately classified IBD status of individual patients during disease quiescence, suggesting that microbial dysbiosis in IBD may be an underlying disorder independent of disease activity. Microbial modules thus provide an integrative view of microbial ecology relevant to IBD.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
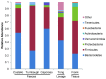
Figure 1. Phylum level microbial compositions of faeces, lavage and tissue samples.
Biospeciemens from faeces (Costello [45], Turnbaugh [12] and Caporaso [11]), lavage samples (Tong) and tissue samples (Frank [21]) were compared. Only predominant phyla with relative abundances higher than 0.1% in Tong dataset were depicted in the bar graph, and the phyla with low abundances were grouped together. For Costello and Caporaso datasets, only the fractions of intestinal microbiota were shown here.
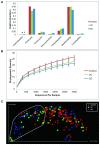
Figure 2. Shifts of mucosal microbial composition in IBD patients in remission.
(A) The change of relative abundance between disease states at phylum level. *: P < 0.05 compared to control, ANOVA. (B) Phylogenetic diversity curves for the microbiota from lavage samples. Mean ± 95% CI was shown. (C) Communities clustered using PCoA of the unweighted UniFrac distance matrix. Each point corresponds to a sample colored by disease phenotype. The dotted line indicated the cluster of samples enriched for IBD subjects.
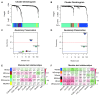
Figure 3. Identification of preserved functional microbial communities (FMCs) associated with disease phenotype across studies.
Hierarchical clustering dendrograms of genera based on microbial co-occurrence network using the Tong dataset (A) and the Frank dataset (B) are shown. In the dendrograms, each color represents one FMC, and each branch represents one genus. The Z-summary statistic plots (y-axis) as a function of the module size are shown for the Tong dataset (C) and the Frank dataset (D). Each point represents a module labeled by color. The dashed blue and red lines indicate the thresholds Z = 2 and Z = 10, respectively. FMC-trait correlations and P values of the Tong dataset (E) and the Frank dataset (F). Each cell reports the correlation coefficient (and P value) derived from correlating FMC eigenvectors (rows) to traits (columns). The table is color-coded by correlation according to the color legend. Collection site: University of California Los Angeles or Cedars Sinai Medical Center; Colon region: 5 anatomical regions coded from 0 to 5, which are cecum, ascending colon, transverse colon, descending colon and rectum.
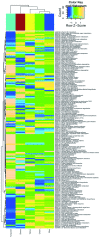
Figure 4. Variations of KEGG metabolic pathways in the functional microbial communities.
The heatmap shows the functional profiles of FMCs (columns) based on the relative abundance of KEGG metabolic pathways (rows) after z score transformation. The color bar on top shows module membership. The dendrograms show the hierarchical clustering of columns and rows respectively using Euclidean distance.
References
- Elson CO, Cong Y, McCracken VJ, Dimmitt RA, Lorenz RG et al. (2005) Experimental models of inflammatory bowel disease reveal innate, adaptive, and regulatory mechanisms of host dialogue with the microbiota. Immunol Rev 206: 260-276. doi: 10.1111/j.0105-2896.2005.00291.x. PubMed: 16048554. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- UL1TR000124/TR/NCATS NIH HHS/United States
- U01 DK062413/DK/NIDDK NIH HHS/United States
- HG005964/HG/NHGRI NIH HHS/United States
- R21 DK084554/DK/NIDDK NIH HHS/United States
- UL1 TR000124/TR/NCATS NIH HHS/United States
- P01DK046763/DK/NIDDK NIH HHS/United States
- DK46763/DK/NIDDK NIH HHS/United States
- R21 HG005964/HG/NHGRI NIH HHS/United States
- R01 AI078885/AI/NIAID NIH HHS/United States
- P01 DK046763/DK/NIDDK NIH HHS/United States
- AI078885/AI/NIAID NIH HHS/United States
- DK062413/DK/NIDDK NIH HHS/United States
- DK084554/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical