Peroxisomes take shape - PubMed (original) (raw)
Review
Peroxisomes take shape
Jennifer J Smith et al. Nat Rev Mol Cell Biol. 2013 Dec.
Abstract
Peroxisomes carry out various oxidative reactions that are tightly regulated to adapt to the changing needs of the cell and varying external environments. Accordingly, they are remarkably fluid and can change dramatically in abundance, size, shape and content in response to numerous cues. These dynamics are controlled by multiple aspects of peroxisome biogenesis that are coordinately regulated with each other and with other cellular processes. Ongoing studies are deciphering the diverse molecular mechanisms that underlie biogenesis and how they cooperate to dynamically control peroxisome utility. These important challenges should lead to an understanding of peroxisome dynamics that can be capitalized upon for bioengineering and the development of therapies to improve human health.
Conflict of interest statement
Competing interests statement
The authors declare no competing interests.
Figures
Figure 1. Peroxisome dynamics
a,b | Immunofluorescence microscopy images of COS-7 cells (African green monkey kidney cells) stained with an antibody against peroxisomal targeting signal 1 (PTS1) (anti-SKL). Different cells in the same culture have different peroxisome morphologies; one cell has spherical peroxisomes (part a), whereas the other cell has both elongated peroxisomes and rows of spherical peroxisomes that are likely to be derived from fission of tubular peroxisomes (part b). c,d | Electron micrographs of the yeast Saccharomyces cerevisiae in the absence (part c) and presence (part d) of fatty acids showing proliferation of peroxisomes (indicated by arrows) in response to the stimulus. e,f | Electron micrographs showing peroxisomes in the yeast Hansenula polymorpha during the development of aggregate-containing peroxisomes (indicated by arrows). The strain agt1_-Cat_mut has blocked autophagic degradation and expresses a mutant version of catalase that form the aggregates, which are sequestered (part e) and separated from the mother organelle by asymmetric peroxisome fission (part f). g | Electron micrograph of Trypansoma brucei showing a class of peroxisomes called glycosomes (indicated by arrows). The images in parts e and f are reproduced, with permission, from REF. © (2013) Landes Bioscience. The image in part g courtesy of Sanjiban Banerjee and Marilyn Parsons, Seattle Biomedical Research Institute, Seattle, Washington, USA.
Figure 2. Direct targeting of proteins to peroxisomes
a | Import of matrix proteins into peroxisomes. Import is shown for a peroxisomal targeting signal 1 (PTS1)-containing cargo (c) imported as a multimer into peroxisomes in yeast. The PTS1 receptor Pex5 interacts with the PTS1 of cargo in the cytoplasm, docks at the docking complex (pink) and is integrated into the membrane to form the transport channel with Pex14. Cargo is released from Pex5 and imported into peroxisomes, and Pex5 is ubiquitylated (not shown) and extracted from the membrane in an ATP-dependent manner by the exportomer (shown in purple). Monoubiquitylation of Pex5 by ubiquitin-conjugating and ligase enzymes, Pex4 and Pex12, respectively, enables Pex5 recycling back to the cytosol for another round of import, whereas polyubiquitylation (by the ubiquitin-conjugating and ligase enzymes, Ubc4 and Pex2, respectively) directs Pex5 to the proteasome for degradation. Pex5 is shown as a tetramer in the cytosol reflecting the mechanistic details of cargo binding and release in Pichia pastoris; Pex5 functions as a tetramer in the cytosol, and as a dimer or heterooligomer (with Pex8) in the peroxisomal membrane. These states seem to be redox-regulated, which promotes cargo binding in the cytoplasm and release in the reducing environment of the peroxisomal lumen. Note that there may be different oligomeric states of PEX5 and functions of each in different organisms. Although most components of the import machinery are evolutionarily conserved, there are notable exceptions, including the existence of functional homologues of PEX26 in Saccharomyces cerevisiae (Pex15), and PEX17 in Neurospora crassa (Pex33). In addition, several peroxins, including Pex22, Pex8, Pex4 and Pex17, have not been identified in higher eukaryotes (TABLE 2). b | Import of membrane proteins into peroxisomes. In the cytoplasm, the peroxisomal membrane protein (PMP) is targeted directly to peroxisomes through the interaction of its membrane PTS (mPTS) with the shuttling receptor Pex19 in the cytoplasm, and through docking of this complex to Pex3 (or PEX16 in mammalian cells) at the peroxisomal membrane. Pex19 then mediates the assembly of PMPs into complexes (shown in blue). PMPs can also be targeted to peroxisomes by insertion into the endoplasmic reticulum (ER) membrane followed by vesicular transport to peroxisomes (FIG. 3).
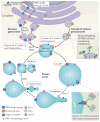
Figure 3. Peroxisomes can form through two pathways
Details of peroxisome formation by de novo generation, and by growth and division are shown for yeast. Peroxisomes are formed de novo from the endoplasmic reticulum (ER) through budding and pair-wise heterotypic fusion of two vesicle types, V1 and V2 (left). This mechanism separates RING finger and docking components of the import complex into different vesicles, which are not import competent until after fusion and assembly of a complete and functional import complex. Peroxin 1 (Pex1) and Pex6 are found in separate preperoxisomal vesicles and are necessary for the heterotypic fusion of V1 and V2 vesicles and the formation of mature, import-competent peroxisomes. Mature peroxisomes can multiply by growth, with proteins and membranes from the ER (via V3 vesicles; right), and fission, mediated by Pex11. Although all preperoxisomal vesicles characterized contain Pex3 (which is required for egression from the ER), V3 vesicles are distinct from V1 and V2 vesicles because they can fuse with mature peroxisomes. A fission cycle begins with membrane remodelling and elongation mediated by Pex11. The elongated extension grows and acquires DRP (dynamin-related protein)-interacting proteins (including the fission protein Fis1 (REFS 88,93,95)). The membrane becomes constricted by an unknown mechanism, and DRPs (Dnm1 and vacuolar sorting protein 1 (Vsp1) in yeast or dynamin-like protein DLP1 in mammalian cells), which are recruited from the cytosol by DRP-interacting proteins, facilitate membrane fission to generate new peroxisomes. Note that for simplicity, not all peroxins are shown.
Similar articles
- Peroxisome biogenesis.
Purdue PE, Lazarow PB. Purdue PE, et al. Annu Rev Cell Dev Biol. 2001;17:701-52. doi: 10.1146/annurev.cellbio.17.1.701. Annu Rev Cell Dev Biol. 2001. PMID: 11687502 Review. - Peroxisome morphology in pathology.
Ribeiro D, Castro I, Fahimi HD, Schrader M. Ribeiro D, et al. Histol Histopathol. 2012 Jun;27(6):661-76. doi: 10.14670/HH-27.661. Histol Histopathol. 2012. PMID: 22473689 Review. - The peroxin Pex34p functions with the Pex11 family of peroxisomal divisional proteins to regulate the peroxisome population in yeast.
Tower RJ, Fagarasanu A, Aitchison JD, Rachubinski RA. Tower RJ, et al. Mol Biol Cell. 2011 May 15;22(10):1727-38. doi: 10.1091/mbc.E11-01-0084. Epub 2011 Mar 25. Mol Biol Cell. 2011. PMID: 21441307 Free PMC article. - Peroxisome biogenesis: advances and conundrums.
Lazarow PB. Lazarow PB. Curr Opin Cell Biol. 2003 Aug;15(4):489-97. doi: 10.1016/s0955-0674(03)00082-6. Curr Opin Cell Biol. 2003. PMID: 12892791 Review. - [Structural, functional and genetic aspects of peroxisome biogenesis].
Kurbatova EM, Dutova TA, Trotsenko IuA. Kurbatova EM, et al. Genetika. 2005 Feb;41(2):149-65. Genetika. 2005. PMID: 15810604 Review. Russian.
Cited by
- A synthetic method to assay polycystin channel biophysics.
Larmore M, Esarte Palomero O, Kamat N, DeCaen PG. Larmore M, et al. Elife. 2024 Oct 28;13:RP98534. doi: 10.7554/eLife.98534. Elife. 2024. PMID: 39466685 Free PMC article. - OrgaMapper: a robust and easy-to-use workflow for analyzing organelle positioning.
Schmied C, Ebner M, Samsó P, Van Der Veen R, Haucke V, Lehmann M. Schmied C, et al. BMC Biol. 2024 Sep 30;22(1):220. doi: 10.1186/s12915-024-02015-8. BMC Biol. 2024. PMID: 39343900 Free PMC article. - STED super-resolution microscopy unveils the dynamics of Atg30 on yeast Pex3-labeled peroxisomes.
de Lange EMF, Mol FN, van der Klei IJ, Vlijm R. de Lange EMF, et al. iScience. 2024 Jul 8;27(8):110481. doi: 10.1016/j.isci.2024.110481. eCollection 2024 Aug 16. iScience. 2024. PMID: 39156652 Free PMC article. - Sex-Specific Dysregulation of Placental Lipid Metabolism in Preeclampsia.
Mishra JS, Zhao H, Zheng J, Kumar S. Mishra JS, et al. Obstet Gynecol Res. 2024;7(3):49-58. doi: 10.26502/ogr0159. Epub 2024 Jul 23. Obstet Gynecol Res. 2024. PMID: 39131546 Free PMC article. - Peroxisomal cholesterol metabolism regulates yap-signaling, which maintains intestinal epithelial barrier function and is altered in Crohn's disease.
Pinelli M, Makdissi S, Scur M, Parsons BD, Baker K, Otley A, MacIntyre B, Nguyen HD, Kim PK, Stadnyk AW, Di Cara F. Pinelli M, et al. Cell Death Dis. 2024 Jul 28;15(7):536. doi: 10.1038/s41419-024-06925-x. Cell Death Dis. 2024. PMID: 39069546 Free PMC article.
References
- Islinger M, Grille S, Fahimi HD, Schrader M. The peroxisome: an update on mysteries. Histochem Cell Biol. 2012;137:547–574. - PubMed
- Pieuchot L, Jedd G. Peroxisome assembly and functional diversity in eukaryotic microorganisms. Annu Rev Microbiol. 2012;66:237–263. - PubMed
- Kunau WH. Peroxisome biogenesis: end of the debate. Curr Biol. 2005;15:R774–R776. - PubMed
- Liu F, Lu Y, Pieuchot L, Dhavale T, Jedd G. Import oligomers induce positive feedback to promote peroxisome differentiation and control organelle abundance. Dev Cell. 2011;21:457–468. Elucidates a cellular mechanism for generating two distinct subpopulations of peroxisomes in the same cell. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM075152/GM/NIGMS NIH HHS/United States
- U01GM098256/GM/NIGMS NIH HHS/United States
- U54GM103511/GM/NIGMS NIH HHS/United States
- R01 GM075152/GM/NIGMS NIH HHS/United States
- U01 GM098256/GM/NIGMS NIH HHS/United States
- U54 GM103511/GM/NIGMS NIH HHS/United States
- P50 GM076547/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources