GnRH-(1-5) transactivates EGFR in Ishikawa human endometrial cells via an orphan G protein-coupled receptor - PubMed (original) (raw)
GnRH-(1-5) transactivates EGFR in Ishikawa human endometrial cells via an orphan G protein-coupled receptor
Madelaine Cho-Clark et al. Mol Endocrinol. 2014 Jan.
Abstract
The decapeptide GnRH is known for its central role in the regulation of the hypothalamo-pituitary-gonadal axis. In addition, it is also known to have local effects within peripheral tissues. The zinc metalloendopeptidase, EC 3.4.24.15 (EP24.15), can cleave GnRH at the Tyr(5)-Gly(6) bond to form the pentapeptide, GnRH-(1-5). The central and peripheral effect of GnRH-(1-5) is different from its parent peptide, GnRH. In the current study, we examined the effect of GnRH-(1-5) on epidermal growth factor receptor (EGFR) phosphorylation and cellular migration. Using the Ishikawa cell line as a model of endometrial cancer, we demonstrate that GnRH-(1-5) stimulates epidermal growth factor release, increases the phosphorylation of EGFR (P < .05) at three tyrosine sites (992, 1045, 1068), and promotes cellular migration. In addition, we also demonstrate that these actions of GnRH-(1-5) are mediated by the orphan G protein-coupled receptor 101 (GPR101). Down-regulation of GPR101 expression blocked the GnRH-(1-5)-mediated release of epidermal growth factor and the subsequent phosphorylation of EGFR and cellular migration. These results suggest that GPR101 is a critical requirement for GnRH-(1-5) transactivation of EGFR in Ishikawa cells.
Figures
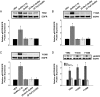
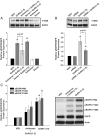
Figure 1.
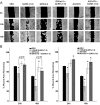
Effect of GnRH-(1–5) on EGFR phosphorylation. Ishikawa cells were treated with 100 nM GnRH-(1–5) for 5 minutes and the phosphorylation of EGFR was measured by Western blot. GnRH-(1–5) increased the phosphorylation of EGFR at sites (A) Y992, (B) Y1045, and (C) Y1068. Conversely, treatment with GnRH or GnRH agonists had no effect on pEGFR levels at any site. (D) Cells with scrambled GnRH-(1–5) (Sc GnRH-[1–5]) exhibited no changes in pEGFR levels relative to VEH. Bars, mean ± SEM (n = 6). *, P < .05 vs VEH.
Figure 2.
Effect of the GnRHR antagonist, Antide, GnRH, and D-Ser6-GnRH on the GnRH-(1–5)-mediated EGFR phosphorylation. (A) Treatment with 100 nM Antide did not alter the ability of 100 nM GnRH-(1–5) (5 min) to increase the levels of pEGFR at any site examined. (B) Treatment with GnRH or D-Ser6-GnRH′′ with (D-Ser6)-GnRH did not have an effect on EGFR phosphorylation. Bars, mean ± SEM (n = 6). *, P < .05 vs VEH.
Figure 3.
Effect of the G protein antagonist, GPAnt-2, or the EGFR antagonist, AG1478, on the GnRH-(1–5)-mediated EGFR phosphorylation. (A) Ishikawa cells treated with GPAnt2 (450 nM) reversed the effect of GnRH-(1–5) (100 nM for 5 min) to increase the phosphorylation of EGFR at sites Y992, Y1045, and Y1068. (B) Blocking EGFR activation with AG1478 (100 nM) inhibited the ability of GnRH-(1–5) to increase pEGFR levels at all sites examined. Bars, SEM (n = 6). *, P < .05 vs VEH.
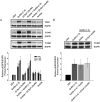
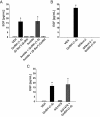
Figure 4.
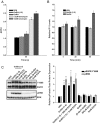
Characterization of the putative GnRH-(1–5) receptor, GPR101, in Ishikawa cells. This is an extension of previous studies that has identified orphan receptors that bind to GnRH-(1–5) (38). (A) Expression of GPR101, GPR119, GPR148, and GPR173 visualized by agarose gel electrophoresis using primers that amplify cDNA from Ishikawa cells. The sequence specificity was confirmed by DNA sequencing. The present study focused on GPR101 due to its previous link to cancer (41). (B) The human GPR101 gene is located on chromosome X site q26.3. The putative mRNA structure consists of a large exon containing the GPR101 coding region. (C) Binding curve using the PathHunter CHO-K1 GPR101 Orphan GPCR β-arrestin high-expression cell line. GnRH-(1–5) activates GPR101 in a dose-dependent mechanism, whereas the GnRH agonist (D-Ser6)-GnRH had no effect. The EC50 of GnRH-(1–5) binding GPR101 in this cell platform is ∼7 nM. (D) We assessed the efficacy of silencing GPR101 expression using antisense oligonucleotide. Ishikawa cells treated with antisense oligonucleotide targeting GPR101 expression reduced the levels of GPR101 mRNA measured by qPCR. As controls, cells were treated with the sense strand or the transfection reagent (OF; oligofectamine) alone with no changes in GPR101 expression levels. Bars, mean ± SEM (n = 4). *, P < .05 vs VEH. (E) To complement the antisense studies, siRNA targeting GPR101 expression was also used to silence GPR101 expression. Treatment with siRNA specifically targeting GPR101 reduced GPR101 mRNA levels by 70%, whereas no significant changes were observed when nontargeting (NT) siRNA was used. Bars, mean ± SEM (n = 3). *, P < .05 vs VEH.
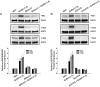
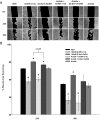
Figure 5.
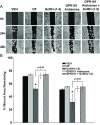
Effect of down-regulating GPR101 expression on the GnRH-(1–5)-mediated EGFR phosphorylation. (A) Ishikawa cells treated with antisense oligonucleotides targeting GPR101 expression had significantly less EGFR phosphorylation when treated with GnRH-(1–5) for 5 minutes. In contrast, cells treated with sense oligonucleotides did not block the ability of GnRH-(1–5) to increase pEGFR (Y1068) levels. Bars, mean ± SEM (n = 4). *, P < .05 vs VEH. (B) Cells were transfected with siRNA specific to GPR101 and EGFR phosphorylation was measured. The effect of GnRH-(1–5) to increase pEGFR levels was reversed only with siRNA targeting GPR101 expression. Bars, mean ± SEM (n = 3). *, P < .05 vs VEH. (C) Moreover, blocking GPR101 expression using an antisense approach also blocked the effect of GnRH-(1–5) on the phosphorylation of EGFR at the sites Y992 and Y1045 complementing the siRNA experiments. Bars, mean ± SEM (n = 4). *, P < .05 vs VEH.
Figure 6.
Effect of GnRH-(1–5) on EGF release in Ishikawa cells. (A) 100 nM GnRH-(1–5) treatment for 5 minutes robustly induced the release of EGF in Ishikawa cells, an effect that was not blocked by Antide. (D-Ser6)-GnRH treatment (100 nM) did not have an effect on EGF release. (B) GnRH-(1–5) stimulation of EGF release was dependent on G protein-coupling because treatment with GPAnt-2 reversed this effect. (C) Treatment with an EGFR kinase inhibitor, AG1478, did not alter the GnRH-(1–5)-induced stimulation of EGF release. Bars, mean ± SEM (n = 6) for all experiments. *, P < .05 vs VEH.
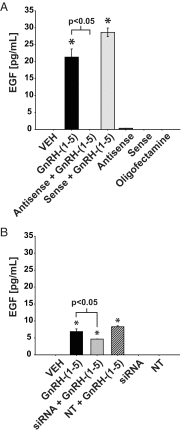
Figure 7.
Effect of silencing GPR101 expression on the GnRH-(1–5)-mediated EGF release. (A) Ishikawa cells treated with 100 nM GnRH-(1–5) for 5 minutes had a significant release in EGF. This increase in EGF was blocked by GPR101 antisense oligonucleotide treatment and not by sense oligonucleotide treatment. (B) siRNA-mediated down-regulation of GPR101 significantly attenuated the ability of GnRH-(1–5) to stimulate release. Treatment with nontargeting siRNA had no effect on GnRH-(1–5) action or basal EGF release. Bars, mean ± SEM (n = 6) for all experiments. *, P < .05 vs VEH.
Figure 8.
Effect of GnRH-(1–5) on cellular migration in Ishikawa cells. The ability of GnRH-(1–5) to regulate cellular migration was examined using a wound closure assay. Ishikawa cells treated with 100 nM GnRH-(1–5) increased migration relative to VEH treatment at 24 and 48 hours, whereas 100 nM GnRH decreased migration. Antide blocked only the effect of GnRH to decrease migration but did not block the migratory effect of GnRH-(1–5). (A, upper panel) The representative photomicrographs of each treatment in the wound closure assay. (B, lower panel) The quantification of the wound closure assay (n = 4). Bars, mean ± SEM (n = 4). *, P < .05 vs VEH. Scale bar, 250 μm.
Figure 9.
Effect of GPAnt-2 or AG1478 on the GnRH-(1–5) regulation of cell migration. To determine whether the G protein antagonist (GPAnt-2) can block the stimulatory effect of GnRH-(1–5) on migration, cells were treated with 450 nM GPAnt-2, which significantly attenuated the effect of GnRH-(1–5) at 24 and 48 hours. Similarly, treatment with the EGFR antagonist, AG1478, reversed the GnRH-(1–5)-mediated increase in cellular migration at 24 and 48 hours. (A, upper panel) The representative photomicrographs of each treatment in the wound closure assay. (B, lower panel) The quantification of the wound closure assay (n = 4) for GPAnt2 (B, left panel) and for AG1478 (B, right panel). Bars, mean ± SEM (n = 4). *, P < .05 vs VEH. Scale bar, 250 μm.
Figure 10.
Effect of silencing GPR101 expression on the GnRH-(1–5) regulation of cell migration. Ishikawa cells were transfected with antisense oligonucleotides targeting GPR101 expression and the ability of GnRH-(1–5) to stimulate wound closure was measured. Cells treated with 100 nM GnRH-(1–5) in the presence or absence of sense oligonucleotides significantly increased migration, whereas treatment with the antisense oligonucleotides blocked the GnRH-(1–5) effect on migration. (A, upper panel) Representative photomicrographs of treatments. (B, lower panel) Quantification of the wound closure assay. Bars, mean ± SEM (n = 4). *, P < .05 vs VEH. Scale bar, 250 μm.
Figure 11.
Effect of Batimastat on EGFR and ERK phosphorylation and EGF release. (A) Ishikawa cells preincubated with a MMP inhibitor, Batimastat, inhibited the GnRH-(1–5)-mediated EGFR and ERK phosphorylation. (B) Batimastast also blocked the GnRH-(1–5)-induced EGF release. Bars, mean ± SEM (n = 4). *, P < .05 vs VEH.
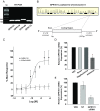
Figure 12.
Effect of GnRH-(1–5) on calcium mobilization. (A) GnRH-(1–5) treatment did not increase intracellular calcium levels, whereas the positive controls, GnRH and ionomycin, increased these levels compared with VEH. (B) Similarly, GnRH-(1–5) had no effect on IP1 accumulation compared with VEH, whereas GnRH demonstrated an increase in intracellular IP1 levels over time. (C) Chelating extracellular calcium with EGTA in cells treated with GnRH-(1–5) did not inhibit the GnRH-(1–5)-mediated EGFR phosphorylation. Bars, mean ± SEM (n = 4). *, P < .05 vs VEH.
Figure 13.
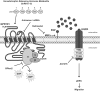
Mechanism of GnRH-(1–5) action in Ishikawa cells. The metabolite GnRH-(1–5) binds GPR101 to induce EGF release in a G protein-dependent mechanism. An increase in EGF release, likely via the cleavage of membrane-bound EGF, induces the phosphorylation of EGFR at the residues Y992, Y1045, and Y1068. Activation of the EGFR pathway in turn initiates signaling cascades important in the promotion of cell migration.
Similar articles
- GnRH-(1-5) activates matrix metallopeptidase-9 to release epidermal growth factor and promote cellular invasion.
Cho-Clark M, Larco DO, Zahn BR, Mani SK, Wu TJ. Cho-Clark M, et al. Mol Cell Endocrinol. 2015 Nov 5;415:114-25. doi: 10.1016/j.mce.2015.08.010. Epub 2015 Aug 12. Mol Cell Endocrinol. 2015. PMID: 26277400 - Matrix metalloproteinases 2 and 9 mediate epidermal growth factor receptor transactivation by gonadotropin-releasing hormone.
Roelle S, Grosse R, Aigner A, Krell HW, Czubayko F, Gudermann T. Roelle S, et al. J Biol Chem. 2003 Nov 21;278(47):47307-18. doi: 10.1074/jbc.M304377200. Epub 2003 Sep 8. J Biol Chem. 2003. PMID: 12963732 - The role of GnRH metabolite, GnRH-(1-5), in endometrial cancer.
Cho-Clark MJ, Watkins A, Wu TJ. Cho-Clark MJ, et al. Front Endocrinol (Lausanne). 2023 Apr 14;14:1183278. doi: 10.3389/fendo.2023.1183278. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 37124730 Free PMC article. Review. - Intracellular signaling pathways mediated by the gonadotropin-releasing hormone (GnRH) receptor.
Kraus S, Naor Z, Seger R. Kraus S, et al. Arch Med Res. 2001 Nov-Dec;32(6):499-509. doi: 10.1016/s0188-4409(01)00331-9. Arch Med Res. 2001. PMID: 11750725 Review.
Cited by
- Comparative transcriptome analysis between patient and endometrial cancer cell lines to determine common signaling pathways and markers linked to cancer progression.
Cho-Clark MJ, Sukumar G, Vidal NM, Raiciulescu S, Oyola MG, Olsen C, Mariño-Ramírez L, Dalgard CL, Wu TJ. Cho-Clark MJ, et al. Oncotarget. 2021 Dec 21;12(26):2500-2513. doi: 10.18632/oncotarget.28161. eCollection 2021 Dec 21. Oncotarget. 2021. PMID: 34966482 Free PMC article. - Sleep Deprivation Alters the Pituitary Stress Transcriptome in Male and Female Mice.
Oyola MG, Shupe EA, Soltis AR, Sukumar G, Paez-Pereda M, Larco DO, Wilkerson MD, Rothwell S, Dalgard CL, Wu TJ. Oyola MG, et al. Front Endocrinol (Lausanne). 2019 Oct 9;10:676. doi: 10.3389/fendo.2019.00676. eCollection 2019. Front Endocrinol (Lausanne). 2019. PMID: 31649619 Free PMC article. - Transactivation of Epidermal Growth Factor Receptor by G Protein-Coupled Receptors: Recent Progress, Challenges and Future Research.
Wang Z. Wang Z. Int J Mol Sci. 2016 Jan 12;17(1):95. doi: 10.3390/ijms17010095. Int J Mol Sci. 2016. PMID: 26771606 Free PMC article. Review. - GHRH excess and blockade in X-LAG syndrome.
Daly AF, Lysy PA, Desfilles C, Rostomyan L, Mohamed A, Caberg JH, Raverot V, Castermans E, Marbaix E, Maiter D, Brunelle C, Trivellin G, Stratakis CA, Bours V, Raftopoulos C, Beauloye V, Barlier A, Beckers A. Daly AF, et al. Endocr Relat Cancer. 2016 Mar;23(3):161-70. doi: 10.1530/ERC-15-0478. Epub 2015 Dec 15. Endocr Relat Cancer. 2016. PMID: 26671997 Free PMC article. - Screening for GPR101 defects in pediatric pituitary corticotropinomas.
Trivellin G, Correa RR, Batsis M, Faucz FR, Chittiboina P, Bjelobaba I, Larco DO, Quezado M, Daly AF, Stojilkovic SS, Wu TJ, Beckers A, Lodish M, Stratakis CA. Trivellin G, et al. Endocr Relat Cancer. 2016 Jun 1;23(5):357-365. doi: 10.1530/ERC-16-0091. Endocr Relat Cancer. 2016. PMID: 26962002 Free PMC article.
References
- Matsuo H, Baba Y, Nair RM, Arimura A, Schally AV. Structure of the porcine LH- and FSH-releasing hormone. I. The proposed amino acid sequence. Biochem Biophys Res Commun. 1971;43:1334–1339. - PubMed
- Tsai PS. Gonadotropin-releasing hormone in invertebrates: structure, function, and evolution. Gen Comp Endocrinol. 2006;148(1):48–53. - PubMed
- Tsai PS, Maldonado TA, Lunden JB. Localization of gonadotropin-releasing hormone in the central nervous system and a peripheral chemosensory organ of Aplysia californica. Gen Comp Endocrinol. 2003;130(1):20–28. - PubMed
- Srkalovic G, Wittliff JL, Schally AV. Detection and partial characterization of receptors for [D-Trp6]-luteinizing hormone-releasing hormone and epidermal growth factor in human endometrial carcinoma. Cancer Res. 1990;50(6):1841–1846. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
This work was supported by The National Science Foundation (IOB-0544150 and IOS-1052288) (to T.J.W.), Department of Defense Grant (RO85FN) (to T.J.W.), and Henry M. Jackson Foundation Graduate Fellowship (to D.O.L.).
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous