Genomics-enabled discovery of phosphonate natural products and their biosynthetic pathways - PubMed (original) (raw)
Review
Genomics-enabled discovery of phosphonate natural products and their biosynthetic pathways
Kou-San Ju et al. J Ind Microbiol Biotechnol. 2014 Feb.
Abstract
Phosphonate natural products have proven to be a rich source of useful pharmaceutical, agricultural, and biotechnology products, whereas study of their biosynthetic pathways has revealed numerous intriguing enzymes that catalyze unprecedented biochemistry. Here we review the history of phosphonate natural product discovery, highlighting technological advances that have played a key role in the recent advances in their discovery. Central to these developments has been the application of genomics, which allowed discovery and development of a global phosphonate metabolic framework to guide research efforts. This framework suggests that the future of phosphonate natural products remains bright, with many new compounds and pathways yet to be discovered.
Figures
Figure 1. Timeline of the discovered phosphonate natural products since 1959
The biosynthetic genes for several compounds have been discovered (blue). Genome mining has thus far yielded the discovery of two new natural products (orange). Compounds that have reached clinical testing, been developed into commercial antibiotics, or biotechnological products are highlighted (red asterisks).
Figure 2. Pathways for phosphonate natural product biosynthesis
Genetic and biochemical characterization of phosphonate producing strains have resulted in the identification of five core biosynthetic pathways common to these natural products. PnPy and PnAA are converted into PMM, 2-keto-4-hydroxy-5-phosphonpentanoic acid (KHPTA), 2-HEP, PnAc, or 2-AEP. Dashed arrows indicate pathways newly discovered as a result of genome mining.
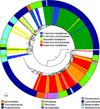
Figure 3. PepM maximum-likelihood tree, nucleotidyl transferases and taxonomy
A maximum-likelihood tree of full-length PepM sequences found in all bacterial genomes in NCBI as of February 2013 is shown. This tree was made using FastTreeMP with the Gamma20 option [50] and altered with the Python library ETE [23]. The inner circle next to the tree is colored based on the presence of an N- or C-terminal nucleotidyltransferase fusion with PepM, along with the presence or absence of a nucleotidyl transferase on a separate ORF. The outer circle shows the phylum level taxonomic designation for each strain. The tree is rooted on the sequence for methylisocitrate lyase from E. coli, indicated with an asterisk.
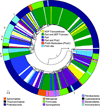
Figure 4. PepM maximum-likelihood tree, early biosynthetic steps, and taxonomy
The phylogenetic tree is the same as shown in Figure 3. Here the inner circle is colored according to the presence of phosphonopyruvate decarboxylase (Ppd), 2-AEP transaminase, phosphonoacetaldehyde reductase (PhpC) and FrbC. The outer circle is the same as in Figure 3 and shows the phylum level taxonomic designation for each strain. Sequences that correspond to known compounds are: 1, bialaphos; 2, cyanophos; 3, dehydrophos; 4, fosfazinomycin; 5, FR-900098; 6, rhizocticin; and 7, fosfomycin. The tree is rooted on the sequence for methylisocitrate lyase from E. coli, indicated with an asterisk.
Similar articles
- Discovery and biosynthesis of phosphonate and phosphinate natural products.
Peck SC, Gao J, van der Donk WA. Peck SC, et al. Methods Enzymol. 2012;516:101-23. doi: 10.1016/B978-0-12-394291-3.00029-0. Methods Enzymol. 2012. PMID: 23034226 - Valinophos Reveals a New Route in Microbial Phosphonate Biosynthesis That Is Broadly Conserved in Nature.
Zhang Y, Chen L, Wilson JA, Cui J, Roodhouse H, Kayrouz C, Pham TM, Ju KS. Zhang Y, et al. J Am Chem Soc. 2022 Jun 8;144(22):9938-9948. doi: 10.1021/jacs.2c02854. Epub 2022 May 26. J Am Chem Soc. 2022. PMID: 35617676 Free PMC article. - A Novel Pathway for Biosynthesis of the Herbicidal Phosphonate Natural Product Phosphonothrixin Is Widespread in Actinobacteria.
Bown L, Hirota R, Goettge MN, Cui J, Krist DT, Zhu L, Giurgiu C, van der Donk WA, Ju KS, Metcalf WW. Bown L, et al. J Bacteriol. 2023 May 25;205(5):e0048522. doi: 10.1128/jb.00485-22. Epub 2023 Apr 19. J Bacteriol. 2023. PMID: 37074199 Free PMC article. - Genomics-directed activation of cryptic natural product pathways deciphers codes for biosynthesis and molecular function.
Tsunematsu Y. Tsunematsu Y. J Nat Med. 2021 Mar;75(2):261-274. doi: 10.1007/s11418-020-01466-x. Epub 2020 Dec 3. J Nat Med. 2021. PMID: 33274411 Free PMC article. Review. - Biosynthesis of phosphonic and phosphinic acid natural products.
Metcalf WW, van der Donk WA. Metcalf WW, et al. Annu Rev Biochem. 2009;78:65-94. doi: 10.1146/annurev.biochem.78.091707.100215. Annu Rev Biochem. 2009. PMID: 19489722 Free PMC article. Review.
Cited by
- Discovery of megapolipeptins by genome mining of a Burkholderiales bacteria collection.
Paulo BS, Recchia MJJ, Lee S, Fergusson CH, Romanowski SB, Hernandez A, Krull N, Liu DY, Cavanagh H, Bos A, Gray CA, Murphy BT, Linington RG, Eustaquio AS. Paulo BS, et al. Chem Sci. 2024 Sep 13;15(40):16567-81. doi: 10.1039/d4sc03594a. Online ahead of print. Chem Sci. 2024. PMID: 39309087 Free PMC article. - Global and seasonal variation of marine phosphonate metabolism.
Lockwood S, Greening C, Baltar F, Morales SE. Lockwood S, et al. ISME J. 2022 Sep;16(9):2198-2212. doi: 10.1038/s41396-022-01266-z. Epub 2022 Jun 23. ISME J. 2022. PMID: 35739297 Free PMC article. - Discovery of phosphonic acid natural products by mining the genomes of 10,000 actinomycetes.
Ju KS, Gao J, Doroghazi JR, Wang KK, Thibodeaux CJ, Li S, Metzger E, Fudala J, Su J, Zhang JK, Lee J, Cioni JP, Evans BS, Hirota R, Labeda DP, van der Donk WA, Metcalf WW. Ju KS, et al. Proc Natl Acad Sci U S A. 2015 Sep 29;112(39):12175-80. doi: 10.1073/pnas.1500873112. Epub 2015 Aug 31. Proc Natl Acad Sci U S A. 2015. PMID: 26324907 Free PMC article. - Deciphering the role of recurrent FAD-dependent enzymes in bacterial phosphonate catabolism.
Zangelmi E, Ruffolo F, Dinhof T, Gerdol M, Malatesta M, Chin JP, Rivetti C, Secchi A, Pallitsch K, Peracchi A. Zangelmi E, et al. iScience. 2023 Oct 4;26(11):108108. doi: 10.1016/j.isci.2023.108108. eCollection 2023 Nov 17. iScience. 2023. PMID: 37876809 Free PMC article. - Transporter characterisation reveals aminoethylphosphonate mineralisation as a key step in the marine phosphorus redox cycle.
Murphy ARJ, Scanlan DJ, Chen Y, Adams NBP, Cadman WA, Bottrill A, Bending G, Hammond JP, Hitchcock A, Wellington EMH, Lidbury IDEA. Murphy ARJ, et al. Nat Commun. 2021 Jul 27;12(1):4554. doi: 10.1038/s41467-021-24646-z. Nat Commun. 2021. PMID: 34315891 Free PMC article.
References
- Baeyer E, Gugel KH, Haegele K, Hagenmaier H, Jessipow S, Koenig WA, Zaehner J. Stofwechselprodukte von Mikroorganismen 98. Phosphinothricin and phosphinothricyle-alanyl-alanin. Helv Chim Acata. 1972;55:224–239. - PubMed
- Bérdy J. Thoughts and facts about antibiotics: Where we are now and where we are heading. The Journal of Antibiotics. 2012;65(8):385–395. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 GM100658/GM/NIGMS NIH HHS/United States
- P01 GM077596/GM/NIGMS NIH HHS/United States
- P01GM077596/GM/NIGMS NIH HHS/United States
- F32GM100658/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources