Insights into mRNA export-linked molecular mechanisms of human disease through a Gle1 structure-function analysis - PubMed (original) (raw)
Review
Insights into mRNA export-linked molecular mechanisms of human disease through a Gle1 structure-function analysis
Andrew W Folkmann et al. Adv Biol Regul. 2014 Jan.
Abstract
A critical step during gene expression is the directional export of nuclear messenger (m)RNA through nuclear pore complexes (NPCs) to the cytoplasm. During export, Gle1 in conjunction with inositol hexakisphosphate (IP6) spatially regulates the activity of the DEAD-box protein Dbp5 at the NPC cytoplasmic face. GLE1 mutations are causally linked to the human diseases lethal congenital contracture syndrome 1 (LCCS-1) and lethal arthrogryposis with anterior horn cell disease (LAAHD). Here, structure prediction and functional analysis provide strong evidence to suggest that the LCCS-1 and LAAHD disease mutations disrupt the function of Gle1 in mRNA export. Strikingly, direct fluorescence microscopy in living cells reveals a dramatic loss of steady-state NPC localization for GFP-gle1 proteins expressed from human gle1 genes harboring LAAHD and LCCS-1 mutations. The potential significance of these residues is further clarified by analyses of sequence and predicted structural conservation. This work offers insights into the perturbed mechanisms underlying human LCCS-1 and LAAHD disease states and emphasizes the potential impact of altered mRNA transport and gene expression in human disease.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures
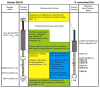
Fig. 1
Schematic depicting functional and structural domains of Human (Left) and S. Cerevisiae (Right) Gle1 proteins is shown. Red dashes indicate the relative position of indicated gle1 alleles. Black arrows mark the location of the conserved IP6-coordinating residues. References cited: (1) Rayala et al., 2004 (2) Folkmann et al., 2013 (3) Alcazar-Roman et al., 2010 (4) Montpetit et al., 2011 (5) Tran et al., 2007 (6) Weirich et al., 2006, Alcazar-Roma et al., 2010 (7) Bolger and Wente, 2011 (8) Murphy and Wente, 1996 (9) Stutz et al., 1997 (10) Strahm et al., 1999 (11) Kendirgi et al., 2005 (12) Kendirgi et al., 2003 (13) Nousianinen et al., 2008.
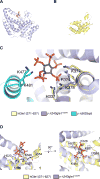
Fig. 2
Identification of putative IP6 coordinating residues in hGle1. (A) Structure of y-Δ243gle1H337R (GRAY) [PDB 3PEU] is shown. (B) A homology model for hGle1(371-627) (YELLOW) was generated based on the crystal structure of y-Δ243gle1H337R [PDB 3PEU] using the structure prediction server Phyre-2 (Keley and Sternberg, 2009). (C–D) This model was constructed by superposing the hGle1(371-627) model onto the y-Δ243gle1H337R molecule within the y-Δ90dbp5/y-Δ243gle1H337R complex (PDB 3PEU). IP6 is rendered as a gray stick molecule with orange phosphate and red oxygen atoms. Nitrogen atoms in Gle1 structures are in blue. (C) Conserved IP6-coordinating polar residues in yGle1 and yDbp5 are labeled. (D) Conserved polar residues in hGle1 are labeled. Methods: Structural analysis and generation of the figure images was done using the program PyMOL (Schrödinger, Inc).
Fig. 3
Charge conservation of the IP6 coordinating residues in Gle1 is observed in some organisms (A–C) Sequence alignment of conserved region of the C-terminal domain of Gle1 from selected fungal and metazoan species. (A) Red box indicates position of yGle1-H337 and hGle1-K486 residues. (B) Red box indicates position of yGle1-R374/K377/K378 and hGle1-H523/K526/K527 residues. (C) Red box indicates position of yGle1-K401 and hGle1-Q554 residues.
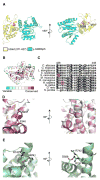
Fig. 4
Conservation of intramolecular salt-bridge in yGle1 and eIF4G. (A) This model was constructed by replacing the y-Δ243gle1H337R molecule within the y-Δ90dbp5/y-Δ243 gle1H337R complex (PDB 3PEU) with the homology model of hGle1(371-627). (B) The results of the analysis of the y-Δ241gle1H337R by the ConSurf server are shown. The Gle1 structure is represented by a ribbon model, colored by the following conservation scale: dark purple residues are the most conserved; white residues are the average on the conservation scale; cyan residues are variable. (C) Sequence alignment of conserved region of the C-terminal domain of Gle1 from selected fungal and metazoan species. Sequences were aligned with ClustalX, shaded with Boxshade 2.1. Red boxes denote the positions of Thr-468 and Val-617 residues in yeast and human Gle1 respectively. (D) Conserved residues in y-Δ243gle1H337R structure are depicted. Dashed line indicated potential hydrogen bond (distance <3Å). (E) Hydrogen bond acceptor and donor residues are indicated in eIF4G structure (PDB 2VSX). Dashed line indicates potential hydrogen bond (distance <3Å). Methods: Structural analysis and generation of the figure images was done using the program PyMOL (Schrödinger, Inc). Multiple sequence alignment analysis was done using the ClustalX, and shaded with Boxshade 3.21
Fig. 5
Conservation of molecular polar contact points in Gle1 and eIF4G. (A–B) The results of the analysis of the (B) y-Δ243gle1H337R [PDB 3PEU] and (C) eIF4G [PDB 2VSX] structures by the ConSurf server are shown. A ribbon model is depicted, colored by the following conservation scale, represents the structures: dark purple residues are the most conserved; white residues are the average on the conservation scale; cyan residues are variable. Conserved polar residues for yGle1 and eIF4G are labeled. (C) Sequence alignment of conserved region of the C-terminal domain of Gle1 from selected fungal and metazoan species. Sequences were aligned with ClustalX, shaded with Boxshade 2.1. Red box denotes the position of the Iso-684 hGle1B residue. Methods: Structural analysis and generation of the figure images was done using the program PyMOL (Schrödinger, Inc). Multiple sequence alignment analysis was done using the ClustalX, and shaded with Boxshade 3.21
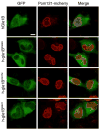
Fig. 6
LAAHD and LCCS-1Het Gle1 proteins have altered steady-state NPC localization. HeLa cells expressing POM121-mCherry and either GFP-hGLE1B, GFP-h-gle1BR569H, GFP-h-gle1BV617M, and GFP-h-gle1BI684T were visualized by direct fluorescent live cell microscopy. Bar, 10 μm. Methods: Hela cells were cultured in complete medium (DMEM, Gibco) supplemented with 10% FBS (Alanta Biologicals) at 37 °C in 5% CO2. Cells were plated in 35mm No. 1.5 glass bottom dishes (Mattek). Transient transfection with indicated plasmids was performed using Fugene6 (Promega) following manufacturer recommendations: POM121-mCherry and pSW1831 (GFP-hGle1B), pSW3971 (GFP-h-gle1BR569H), pSW3972 (GFP-h-gle1BV617M), or pSW3973 (GFP-h-gle1BI684T). All live-cell direct fluorescence microscopy experiments were performed on a confocal microscope (LSM710, Zeiss, 40X/1.1 C-Apochromat water objective).
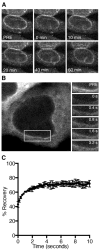
Fig. 7
Gle1 is a stable component of the NPC. (A) HeLa cells expressing GFP-hGLE1B were analyzed by FRAP microscopy. Representative nuclear rim FRAP time series images are shown. Bar, 10 μm. (B) HeLa cells expressing GFP-hDBP5 were analyzed by FRAP microscopy. Representative nuclear rim FRAP time series images are shown. Bar, 10 μm. White box indicates imaging region of interest for FRAP acquisition (C) FRAP recovery curve experimental determined bleached region, fit with a one-phase association model. Error bars represent mean ± standard deviation with n=5 cells. Methods: HeLa cells were cultured and transfected as in Figure 4. FRAP microscopy experiments were performed on HeLa cells co-transfected with POM121-mCherry and either pSW1832, or pSW3253 (GFP-hDBP5). The bleaching region of interest (B-ROI) was set to encompass the nuclear rim. Bleaching was achieved by exciting at 488 nM throughout the entire B-ROI (with a LSM710, Zeiss, 40X/1.1 C-Apochromat water objective). Post-bleaching images were acquired every 10 minutes (GFP-hGle1) or 200ms (GFP-hDbp5).
Fig. 8
Charge conservation of the IP6 coordinating residues in Dbp5 is observed in some organisms. (A) Sequence alignment of the far C-terminal region in Dbp5 from selected fungal and metazoan species. Red box indicates position of yDbp5-K477/K481 residues.
Similar articles
- Survival beyond the perinatal period expands the phenotypes caused by mutations in GLE1.
Said E, Chong JX, Hempel M, Denecke J, Soler P, Strom T, Nickerson DA, Kubisch C; University of Washington Center for Mendelian Genomics; Bamshad MJ, Lessel D. Said E, et al. Am J Med Genet A. 2017 Nov;173(11):3098-3103. doi: 10.1002/ajmg.a.38406. Epub 2017 Sep 8. Am J Med Genet A. 2017. PMID: 28884921 Free PMC article. - Nup42 and IP6 coordinate Gle1 stimulation of Dbp5/DDX19B for mRNA export in yeast and human cells.
Adams RL, Mason AC, Glass L, Aditi, Wente SR. Adams RL, et al. Traffic. 2017 Dec;18(12):776-790. doi: 10.1111/tra.12526. Epub 2017 Oct 16. Traffic. 2017. PMID: 28869701 Free PMC article. - The Dbp5 cycle at the nuclear pore complex during mRNA export I: dbp5 mutants with defects in RNA binding and ATP hydrolysis define key steps for Nup159 and Gle1.
Hodge CA, Tran EJ, Noble KN, Alcazar-Roman AR, Ben-Yishay R, Scarcelli JJ, Folkmann AW, Shav-Tal Y, Wente SR, Cole CN. Hodge CA, et al. Genes Dev. 2011 May 15;25(10):1052-64. doi: 10.1101/gad.2041611. Genes Dev. 2011. PMID: 21576265 Free PMC article. - Dbp5, Gle1-IP6 and Nup159: a working model for mRNP export.
Folkmann AW, Noble KN, Cole CN, Wente SR. Folkmann AW, et al. Nucleus. 2011 Nov-Dec;2(6):540-8. doi: 10.4161/nucl.2.6.17881. Epub 2011 Nov 1. Nucleus. 2011. PMID: 22064466 Free PMC article. Review. - Dbp5 - from nuclear export to translation.
Tieg B, Krebber H. Tieg B, et al. Biochim Biophys Acta. 2013 Aug;1829(8):791-8. doi: 10.1016/j.bbagrm.2012.10.010. Epub 2012 Nov 2. Biochim Biophys Acta. 2013. PMID: 23128325 Review.
Cited by
- Identification of the Novel Nup188-brr7 Allele in a Screen for Cold-Sensitive mRNA Export Mutants in Saccharomyces cerevisiae.
de Bruyn Kops A, Guthrie C. de Bruyn Kops A, et al. G3 (Bethesda). 2018 Aug 30;8(9):2991-3003. doi: 10.1534/g3.118.200447. G3 (Bethesda). 2018. PMID: 30021831 Free PMC article. - Structural and functional analysis of mRNA export regulation by the nuclear pore complex.
Lin DH, Correia AR, Cai SW, Huber FM, Jette CA, Hoelz A. Lin DH, et al. Nat Commun. 2018 Jun 13;9(1):2319. doi: 10.1038/s41467-018-04459-3. Nat Commun. 2018. PMID: 29899397 Free PMC article. - An amyotrophic lateral sclerosis-linked mutation in GLE1 alters the cellular pool of human Gle1 functional isoforms.
Aditi, Glass L, Dawson TR, Wente SR. Aditi, et al. Adv Biol Regul. 2016 Sep;62:25-36. doi: 10.1016/j.jbior.2015.11.001. Epub 2015 Nov 11. Adv Biol Regul. 2016. PMID: 26776475 Free PMC article. - Multiple Export Mechanisms for mRNAs.
Delaleau M, Borden KL. Delaleau M, et al. Cells. 2015 Aug 28;4(3):452-73. doi: 10.3390/cells4030452. Cells. 2015. PMID: 26343730 Free PMC article. Review. - The Nuclear Pore Complex and mRNA Export in Cancer.
Borden KLB. Borden KLB. Cancers (Basel). 2020 Dec 25;13(1):42. doi: 10.3390/cancers13010042. Cancers (Basel). 2020. PMID: 33375634 Free PMC article. Review.
References
- Al-Qattan MM, Shamseldin HE, Alkuraya FS. Familial dorsalization of the skin of the proximal palm and the instep of the sole of the foot. Gene. 2012;500:216–9. - PubMed
- Alcazar-Roman AR, Tran EJ, Guo S, Wente SR. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nature Cell Biology. 2006;8:711–6. - PubMed
- Ballut L, Marchadier B, Baguet A, Tomasetto C, Seraphin B, Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol. 2005;12:861–9. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- T32 CA119925/CA/NCI NIH HHS/United States
- F31NS070431/NS/NINDS NIH HHS/United States
- T32CA119925/CA/NCI NIH HHS/United States
- R01 GM051219/GM/NIGMS NIH HHS/United States
- P30CA068485/CA/NCI NIH HHS/United States
- R37 GM51219/GM/NIGMS NIH HHS/United States
- F31 NS070431/NS/NINDS NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- R37 GM051219/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources