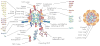Enriching the pore: splendid complexity from humble origins - PubMed (original) (raw)
Review
Enriching the pore: splendid complexity from humble origins
Mark C Field et al. Traffic. 2014 Feb.
Abstract
The nucleus is the defining intracellular organelle of eukaryotic cells and represents a major structural innovation that differentiates the eukaryotic and prokaryotic cellular form. The presence of a nuclear envelope (NE) encapsulating the nucleus necessitates a mechanism for interchange between the contents of the nuclear interior and the cytoplasm, which is mediated via the nuclear pore complex (NPC), a large protein assembly residing in nuclear pores in the NE. Recent advances have begun to map the structure and functions of the NPC in multiple organisms, and to allow reconstruction of some of the evolutionary events that underpin the modern NPC form, highlighting common and differential NPC features across the eukaryotes. Here we discuss some of these advances and the questions being pursued, consider how the evolution of the NPC has been constrained, and finally propose a model for how the NPC evolved.
Keywords: comparative genomics; eukaryogenesis; molecular evolution; nuclear pore complex; nucleus; protocoatomer.
© 2013 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd.
Figures
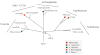
Figure 1. Schematic phylogenetic tree of the eukaryotes
The major taxonomic groupings are shown, and the overall topology corresponds to the most likely based on present data and analytical methods. Supergroups are indicated by bars, and orders are labelled at the various nodes. Colored blobs indicate the positions of specific taxa that are discussed in the article and specified in the key. LECA and FECA are the last and first eukaryotic common ancestors respectively.
Figure 2. The NPC; conserved and non-conserved regions
Panel A: Schematic of the nuclear pore complex color-coded to emphasis distinct structural modules within the structure. The central channel is shown with FG-repeat Nups as noodle-like structures, while the scaffold is shown as globular lozenges. Connections between the NPC core and the lamina are also shown. Several transport cargo systems are also shown. The structure is rotated through 90° at right to shown the organelle from the cytoplasmic face. Panel B: Schematics of the nuclear pore complex color-coded to match Panel A and C with individual Nups listed inside the major structural modules. The nomenclature is according to the human Nups with the exception of the names starting with Sc for Nups restricted to yeasts. The names of the subunits that were likely present in LECA based on comparative genomics (Panel C) are in black. The presence of the Nups with names in gray in LECA is more speculative. Lineage specific Nups are in white. Panel C: Coulson plot representation of nucleoporins across the eukaryotes as revealed by genomics. Well characterized Nups from vertebrates (Hs), yeast (Sc) and their homologs as identified in other eukaryotic lineages are shown as a Coulson plot. The data are based on the analysis of Neumann et al. (36) and our own additional homology searches using the HMMER software and protein alignments of clearly homologous protein sequences of the known Nups of metazoa and/or fungi as queries. Positive hits in other eukaryotic lineages were verified by reverse BLAST or HMMER homology searches. The composition of the NPC of Cryptophytes and Chlorarachniophytes is based on annotation of the NPC subunits in these algae in Curtis et al. (80). The color scheme of the nucleoporin modules is the same as in Panel A and B. Some of the subunits in the kinetoplastid lineage were found by protein-interaction analyses in T. brucei and are assigned as Nups based on similar secondary structure even when low amino-acid sequence similarity with validated Nups from vertebrates and fungi was found (empty circles). Blue symbols indicate experimentally validated Nups.
Figure 2. The NPC; conserved and non-conserved regions
Panel A: Schematic of the nuclear pore complex color-coded to emphasis distinct structural modules within the structure. The central channel is shown with FG-repeat Nups as noodle-like structures, while the scaffold is shown as globular lozenges. Connections between the NPC core and the lamina are also shown. Several transport cargo systems are also shown. The structure is rotated through 90° at right to shown the organelle from the cytoplasmic face. Panel B: Schematics of the nuclear pore complex color-coded to match Panel A and C with individual Nups listed inside the major structural modules. The nomenclature is according to the human Nups with the exception of the names starting with Sc for Nups restricted to yeasts. The names of the subunits that were likely present in LECA based on comparative genomics (Panel C) are in black. The presence of the Nups with names in gray in LECA is more speculative. Lineage specific Nups are in white. Panel C: Coulson plot representation of nucleoporins across the eukaryotes as revealed by genomics. Well characterized Nups from vertebrates (Hs), yeast (Sc) and their homologs as identified in other eukaryotic lineages are shown as a Coulson plot. The data are based on the analysis of Neumann et al. (36) and our own additional homology searches using the HMMER software and protein alignments of clearly homologous protein sequences of the known Nups of metazoa and/or fungi as queries. Positive hits in other eukaryotic lineages were verified by reverse BLAST or HMMER homology searches. The composition of the NPC of Cryptophytes and Chlorarachniophytes is based on annotation of the NPC subunits in these algae in Curtis et al. (80). The color scheme of the nucleoporin modules is the same as in Panel A and B. Some of the subunits in the kinetoplastid lineage were found by protein-interaction analyses in T. brucei and are assigned as Nups based on similar secondary structure even when low amino-acid sequence similarity with validated Nups from vertebrates and fungi was found (empty circles). Blue symbols indicate experimentally validated Nups.
Figure 3. Phylogenetic reconstruction of Mlp nucleoporin evolution
Maximum likelihood phylogenetic tree of the TPR protein family. Broad distribution of these proteins in distinct eukaryotic lineages indicates that it was present in LECA. Vast majority of taxa possess a single gene in their genomes; the gene duplications that led to Mlp1/Mlp2 in S. cerevisiae and Nup211/Alm1 of S. pombe (indicated by red symbols) are two independent events that occurred later in the evolution of fungi. Numbers at nodes are SH-like aLRT values calculated in PhyML 3.1.
Figure 4. Evolution of the NPC, nucleus and the ER
A simple model is proposed for how the nucleus could have evolved from a prokaryotic ancestor that lacked a differentiated cytoplasm. Initially the DNA is untethered to membrane (left), but in some bacterial lineages both the presence of membrane microdomains and DNA tethering is known (second left). Invaginations of membrane, which may have been accompanied by the presence of a primitive protocoatomer complex are envisaged. As these structures become more complex, stable internal membrane have arisen, which may maintain a connection with the plasma membrane (third left). At this point the internal membranes are in essence evolving into a proto-ER, as they may share protein export machinery and other factors with the bacterial plasma membrane. Maintaining these structures within the cytosol requires a coating system at the rim/apex of the membrane tubules or sheets for stability, control of proliferation and possible membrane fission. Once the DNA relocates to these membranes, these membrane act as a bivouac for the DNA, and are essentially transformed into a proto-nucleus (third right). Close proximity of membranes, which would have been fenestrated to facilitate exchange of molecules between the DNA and cytoplasm, requires a coat complex to bring stability to these membrane structures, and which also serves as the beginning of a nuclear pore and NPC; at this stage we suggest that the system would be a non-specific pore lacking a gating function. Later the system becomes more differentiated, and the appearance of FG-repeat nucleoporins serve to introduce gating, specificity and to increase targeting efficiency (second right), until a fully differentiated nucleus has arisen, which finally parts company with the plasma membrane (right). Note that the model ignores many critical aspects of eukaryogenesis, including endosymbiosis and membrane trafficking and many other process, as well as evidence that such states constitute true intermediates. It also assumes that the nucleus arose by a fully autogenous mechanism, which is generally, if not universally, accepted. Schematics of idealised cells are shown at center; with the true nuclear envelope and DNA in blue. The implied configuration of the protocoatomer ancestor leading up to the NPC is shown below; FG-repeat NUPs are shown in green, the nuclear envelope in gray and the NPC core scaffold in blue. The large gray arrow indicates a timebase.
Similar articles
- Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex.
Mans BJ, Anantharaman V, Aravind L, Koonin EV. Mans BJ, et al. Cell Cycle. 2004 Dec;3(12):1612-37. doi: 10.4161/cc.3.12.1316. Epub 2004 Dec 20. Cell Cycle. 2004. PMID: 15611647 - The nuclear pore complex core scaffold and permeability barrier: variations of a common theme.
Hayama R, Rout MP, Fernandez-Martinez J. Hayama R, et al. Curr Opin Cell Biol. 2017 Jun;46:110-118. doi: 10.1016/j.ceb.2017.05.003. Epub 2017 Jun 15. Curr Opin Cell Biol. 2017. PMID: 28624666 Free PMC article. Review. - The nuclear pore complex: a strategic platform for regulating cell signaling.
Gu Y. Gu Y. New Phytol. 2018 Jul;219(1):25-30. doi: 10.1111/nph.14756. Epub 2017 Aug 31. New Phytol. 2018. PMID: 28858378 Review. - Functional architecture of the nuclear pore complex.
Grossman E, Medalia O, Zwerger M. Grossman E, et al. Annu Rev Biophys. 2012;41:557-84. doi: 10.1146/annurev-biophys-050511-102328. Annu Rev Biophys. 2012. PMID: 22577827 Review. - Integrative structure-function mapping of the nucleoporin Nup133 suggests a conserved mechanism for membrane anchoring of the nuclear pore complex.
Kim SJ, Fernandez-Martinez J, Sampathkumar P, Martel A, Matsui T, Tsuruta H, Weiss TM, Shi Y, Markina-Inarrairaegui A, Bonanno JB, Sauder JM, Burley SK, Chait BT, Almo SC, Rout MP, Sali A. Kim SJ, et al. Mol Cell Proteomics. 2014 Nov;13(11):2911-26. doi: 10.1074/mcp.M114.040915. Epub 2014 Aug 19. Mol Cell Proteomics. 2014. PMID: 25139911 Free PMC article.
Cited by
- Extensive Reduction of the Nuclear Pore Complex in Nucleomorphs.
Irwin NAT, Keeling PJ. Irwin NAT, et al. Genome Biol Evol. 2019 Mar 1;11(3):678-687. doi: 10.1093/gbe/evz029. Genome Biol Evol. 2019. PMID: 30715330 Free PMC article. - Nuclear envelope: a new frontier in plant mechanosensing?
Fal K, Asnacios A, Chabouté ME, Hamant O. Fal K, et al. Biophys Rev. 2017 Aug;9(4):389-403. doi: 10.1007/s12551-017-0302-6. Epub 2017 Aug 12. Biophys Rev. 2017. PMID: 28801801 Free PMC article. Review. - A lineage-specific protein network at the trypanosome nuclear envelope.
Butterfield ER, Obado SO, Scutts SR, Zhang W, Chait BT, Rout MP, Field MC. Butterfield ER, et al. Nucleus. 2024 Dec;15(1):2310452. doi: 10.1080/19491034.2024.2310452. Epub 2024 Apr 11. Nucleus. 2024. PMID: 38605598 Free PMC article. - The Inner Nuclear Membrane Protein Src1 Is Required for Stable Post-Mitotic Progression into G1 in Aspergillus nidulans.
Liu HL, Osmani AH, Osmani SA. Liu HL, et al. PLoS One. 2015 Jul 6;10(7):e0132489. doi: 10.1371/journal.pone.0132489. eCollection 2015. PLoS One. 2015. PMID: 26147902 Free PMC article. - The nucleus: keeping it together by keeping it apart.
Lusk CP, King MC. Lusk CP, et al. Curr Opin Cell Biol. 2017 Feb;44:44-50. doi: 10.1016/j.ceb.2017.02.001. Epub 2017 Feb 23. Curr Opin Cell Biol. 2017. PMID: 28236735 Free PMC article. Review.
References
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. 4. New York: Garland Science; 2002.
- Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem. 2011;80:613–643. - PubMed
- Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. - PubMed
- Tetenbaum-Novatt J, Rout MP. The mechanism of nucleocytoplasmic transport through the nuclear pore complex. Cold Spring Harb Symp Quant Biol. 2010;75:567–584. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U54 GM103511/GM/NIGMS NIH HHS/United States
- R21 AI096069/AI/NIAID NIH HHS/United States
- 090007/Z/09/Z/WT_/Wellcome Trust/United Kingdom
- U01 GM098256/GM/NIGMS NIH HHS/United States
- MR/K008749/1/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources