Ets-1 global gene expression profile reveals associations with metabolism and oxidative stress in ovarian and breast cancers - PubMed (original) (raw)
Ets-1 global gene expression profile reveals associations with metabolism and oxidative stress in ovarian and breast cancers
Meghan L Verschoor et al. Cancer Metab. 2013.
Abstract
Background: The Ets-1 proto-oncogene is frequently upregulated in cancer cells, with known involvement in cancer angiogenesis, metastasis, and more recently energy metabolism. In this study we have performed various bioinformatic analyses on existing microarray data to further clarify the role of Ets-1 in ovarian cancer, and validated these results with functional assays.
Methods: Functional pathway analyses were conducted on existing microarray data comparing 2008 and 2008-Ets1 ovarian cancer cells. Methods included over-representation analysis, functional class scoring and pathway topology, and network representations were visualized in Cytoscape. Oxidative stress regulation was examined in ovarian cancer cells by measuring protein expression and enzyme activity of glutathione peroxidases, as well as intracellular reactive oxygen species using dichlorofluorescin fluorescence. A stable Ets-1 knockdown MDA-MB-231 cell line was created using short hairpin RNA, and glycolytic dependence of these cells was measured following treatment with 2-deoxy-D-glucose and Hoechst nuclear staining to determine cell number. High-resolution respirometry was performed to measure changes in basal oxygen flux between MDA-MB-231 cells and MDA-Ets1KD variants.
Results: Enrichments in oxidoreductase activity and various metabolic pathways were observed upon integration of the different analyses, suggesting that Ets-1 is important in their regulation. As oxidative stress is closely associated with these pathways, we functionally validated our observations by showing that Ets-1 overexpression resulted in decreased reactive oxygen species with increased glutathione peroxidase expression and activity, thereby regulating cellular oxidative stress. To extend our findings to another cancer type, we developed an Ets-1 knockdown breast cancer cell model, which displayed decreased glycolytic dependence and increased oxygen consumption following Ets-1 knockdown confirming our earlier findings.
Conclusions: Collectively, this study confirms the important role of Ets-1 in the regulation of cancer energy metabolism in ovarian and breast cancers. Furthermore, Ets-1 is a key regulator of oxidative stress in ovarian cancer cells by mediating alterations in glutathione antioxidant capacity.
Figures
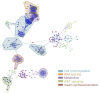
Figure 1
Global functional interaction network. Reactome FI was used to create a functional interaction network, which is divided into modules as defined by node coloring, and further delineated by functional pathway enrichments into outlined, color-coded groups. Major associations among the curated and predicted interactions included cell cycle regulatory, RNA slicing, metabolism, WNT signaling, and insulin-related pathways (FDR <0.01). Network was generated in Cytoscape.
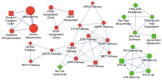
Figure 2
Enrichment map of gene set enrichment analysis. The map shows enriched gene sets in 2008 versus 2008-Ets1 ovarian cancer cells clustered by MCODE to generate sub-networks of the interrelated gene sets. Red nodes indicate enrichment (upregulation) in 2008 cells, green nodes represent enrichment (upregulation) in 2008-Ets1 cells. Node size is representative of the number of enriched genes in the gene set. The largest cluster includes the signaling pathways of ERK5, MAPK, EGF, PDGF, MET and GPCR. Notably, clusters with gene sets involved in mitochondrial metabolism and fatty acid metabolic processes were also identified. Network was generated in Cytoscape.
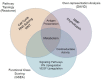
Figure 3
Integrating various bioinformatic analyses. Venn diagram representing the overlapping enrichments from the various bioinformatic pathway analyses (Figures 1 and 2, Table 1) employed on the microarray expression data. The functional interaction network and ontological analyses both included enrichments in antigen presentation, and the ontological analysis shared oxidoreductase activity enrichment with the gene set enrichment analysis. All three analyses found enrichments in various metabolic pathways.
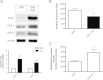
Figure 4
Ets-1 regulated oxidative stress in ovarian cancer cells. (A) The protein expression of Ets1, GPX-1 and GPX-2 were examined via western blot, and normalized to Actin expression by densitometry analysis. Ets-1 overexpressing cells show 3.10-fold and 2.25-fold inductions in GPX-1 and GPX-2 protein levels, respectively. (B) Intracellular ROS levels were measured using the fluorescent CM2-H2DCFDA reagent in ovarian cancer cells. 2008-Ets1 cells contained lower ROS levels than 2008 cells (1233.99 AFU and 1872.73 AFU, respectively). (C) The activity of glutathione peroxidase enzymes was measured using a colorimetric assay, where Ets-1 overexpressing cells were observed to have significantly higher activity than parental cells (7725.66 U/mL/mg and 3944.22 U/mL/mg, respectively).
Figure 5
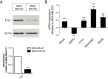
Breast cancer cell model of Ets-1 expression knockdown. MDA-MB-231 breast cancer cells were stably depleted of Ets-1 expression via targeted shRNA knockdown. (A) The Ets-1 knockdown cell line MDA-Ets1KD expresses Ets-1 protein at 22.7% of parental protein levels in MDA-MB-231 cells. (B) Real-time qRT-PCR of the breast cancer Ets-1 expression model. The gene expression of PDHA, CYC1, NDUFAB1 and SDHB were increased in MDA-Ets1KD cells, whereas the expression of G6PD was downregulated.
Figure 6
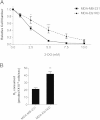
Effect of Ets-1 knockdown on breast cancer cell metabolism. (A) MDA-MB-231 and MDA-Ets1KD cells were treated with various amounts of the glycolytic inhibitor 2-DG, and representative growth curves were generated for each cell line. The 2-DG IC50 was greater in MDA-Ets1KD cells, with an IC50 of 3.94 mM compared to an IC50 of 2.03 mM for MDA-MB-231 cells. (B) Basal oxygen consumption was measured using high-resolution respirometry, and MDA-Est1KD cells were observed to consume significantly more oxygen (41.80 pmol/106 cells/s) than parental MDA-MB-231 cells (21.07 pmol/106 cells/s).
Similar articles
- Ets-1 regulates intracellular glutathione levels: key target for resistant ovarian cancer.
Verschoor ML, Singh G. Verschoor ML, et al. Mol Cancer. 2013 Nov 15;12(1):138. doi: 10.1186/1476-4598-12-138. Mol Cancer. 2013. PMID: 24238102 Free PMC article. - Ets-1 regulates energy metabolism in cancer cells.
Verschoor ML, Wilson LA, Verschoor CP, Singh G. Verschoor ML, et al. PLoS One. 2010 Oct 22;5(10):e13565. doi: 10.1371/journal.pone.0013565. PLoS One. 2010. PMID: 21042593 Free PMC article. - Gene expression profile analysis of an isogenic tumour metastasis model reveals a functional role for oncogene AF1Q in breast cancer metastasis.
Li DQ, Hou YF, Wu J, Chen Y, Lu JS, Di GH, Ou ZL, Shen ZZ, Ding J, Shao ZM. Li DQ, et al. Eur J Cancer. 2006 Dec;42(18):3274-86. doi: 10.1016/j.ejca.2006.07.008. Epub 2006 Sep 18. Eur J Cancer. 2006. PMID: 16979889 - A systematic review of p53 regulation of oxidative stress in skeletal muscle.
Beyfuss K, Hood DA. Beyfuss K, et al. Redox Rep. 2018 Dec;23(1):100-117. doi: 10.1080/13510002.2017.1416773. Epub 2018 Jan 3. Redox Rep. 2018. PMID: 29298131 Free PMC article. Review. - The Ets family contains transcriptional activators and repressors involved in angiogenesis.
Lelièvre E, Lionneton F, Soncin F, Vandenbunder B. Lelièvre E, et al. Int J Biochem Cell Biol. 2001 Apr;33(4):391-407. doi: 10.1016/s1357-2725(01)00025-5. Int J Biochem Cell Biol. 2001. PMID: 11312108 Review.
Cited by
- Allele-specific effect of various dietary fatty acids and ETS1 transcription factor on SCD1 expression.
Tibori K, Zámbó V, Orosz G, Szelényi P, Sarnyai F, Tamási V, Rónai Z, Csala M, Kereszturi É. Tibori K, et al. Sci Rep. 2024 Jan 2;14(1):177. doi: 10.1038/s41598-023-50700-5. Sci Rep. 2024. PMID: 38167845 Free PMC article. - Ets1 facilitates EMT/invasion through Drp1-mediated mitochondrial fragmentation in ovarian cancer.
Ghosh D, Pakhira S, Ghosh DD, Roychoudhury S, Roy SS. Ghosh D, et al. iScience. 2023 Aug 2;26(9):107537. doi: 10.1016/j.isci.2023.107537. eCollection 2023 Sep 15. iScience. 2023. PMID: 37664613 Free PMC article. - Distinct Prognostic and Immunological Roles of ETS1 and ETS2: A Pan-Cancer Analysis.
Ren Y, Chen B, Zhang M. Ren Y, et al. Biomed Res Int. 2023 Jan 31;2023:4343350. doi: 10.1155/2023/4343350. eCollection 2023. Biomed Res Int. 2023. PMID: 36760475 Free PMC article. - Ets1 mediates sorafenib resistance by regulating mitochondrial ROS pathway in hepatocellular carcinoma.
Vishnoi K, Ke R, Viswakarma N, Srivastava P, Kumar S, Das S, Singh SK, Principe DR, Rana A, Rana B. Vishnoi K, et al. Cell Death Dis. 2022 Jul 4;13(7):581. doi: 10.1038/s41419-022-05022-1. Cell Death Dis. 2022. PMID: 35789155 Free PMC article. - The histone demthylase KDM3A protects the myocardium from ischemia/reperfusion injury via promotion of ETS1 expression.
Guo X, Zhang BF, Zhang J, Liu G, Hu Q, Chen J. Guo X, et al. Commun Biol. 2022 Mar 25;5(1):270. doi: 10.1038/s42003-022-03225-y. Commun Biol. 2022. PMID: 35338235 Free PMC article.
References
- Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. - PubMed
- Baldus CD, Liyanarachchi S, Mrozek K, Auer H, Tanner SM, Guimond M, Ruppert AS, Mohamed N, Davuluri RV, Caligiuri MA, Bloomfield CD, de la Chapelle A. Acute myeloid leukemia with complex karyotypes and abnormal chromosome 21: Amplification discloses overexpression of APP, ETS2, and ERG genes. Proc Natl Acad Sci U S A. 2004;101(11):3915–3920. doi: 10.1073/pnas.0400272101. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous