Fecal microbiomes of non-human primates in Western Uganda reveal species-specific communities largely resistant to habitat perturbation - PubMed (original) (raw)
doi: 10.1002/ajp.22238. Epub 2013 Nov 27.
Colin A Chapman, Geoffrey Weny, Alex Tumukunde, David Hyeroba, Kelly Klotz, Avery S Koblings, David N M Mbora, Melissa Cregger, Bryan A White, Steven R Leigh, Tony L Goldberg
Affiliations
- PMID: 24285224
- PMCID: PMC4097101
- DOI: 10.1002/ajp.22238
Fecal microbiomes of non-human primates in Western Uganda reveal species-specific communities largely resistant to habitat perturbation
Aleia I McCord et al. Am J Primatol. 2014 Apr.
Abstract
Primate gastrointestinal microbial communities are becoming increasingly appreciated for their relevance to comparative medicine and conservation, but the factors that structure primate "microbiomes" remain controversial. This study examined a community of primates in Kibale National Park, Uganda, to assess the relative importance of host species and location in structuring gastrointestinal microbiomes. Fecal samples were collected from primates in intact forest and from primates in highly disturbed forest fragments. People and livestock living nearby were also included, as was a geographically distant population of related red colobus in Kenya. A culture-free microbial community fingerprinting technique was used to analyze fecal microbiomes from 124 individual red colobus (Procolobus rufomitratus), 100 individual black-and-white colobus (Colobus guereza), 111 individual red-tailed guenons (Cercopithecus ascanius), 578 human volunteers, and 364 domestic animals, including cattle (Bos indicus and B. indicus × B. taurus crosses), goats (Caprus hircus), sheep (Ovis aries), and pigs (Sus scrofa). Microbiomes sorted strongly by host species, and forest fragmentation did not alter this pattern. Microbiomes of Kenyan red colobus sorted distinctly from microbiomes of Ugandan red colobus, but microbiomes from these two red colobus populations clustered more closely with each other than with any other species. Microbiomes from red colobus and black-and-white colobus were more differentiated than would be predicted by the phylogenetic relatedness of these two species, perhaps reflecting heretofore underappreciated differences in digestive physiology between the species. Within Kibale, social group membership influenced intra-specific variation among microbiomes. However, intra-specific variation was higher among primates in forest fragments than among primates in intact forest, perhaps reflecting the physical separation of fragments. These results suggest that, in this system, species-specific processes such as gastrointestinal physiology strongly structure microbial communities, and that primate microbiomes are relatively resistant to perturbation, even across large geographic distances or in the face of habitat disturbance.
Keywords: Cercopithecinae; Colobinae; Uganda; forest fragmentation; microbiome; non-human primate.
© 2013 Wiley Periodicals, Inc.
Figures
Fig. 1
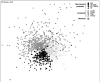
Non-metric multi-dimensional scaling ordination based on Bray–Curtis similarity matrix for fecal microbiomes of three primate species from Kibale National Park (KNP) and surrounding forest fragments: red colobus (RC), black-and-white colobus (BW), and red-tailed guenon (RT). Red colobus from Tana River National Park, Kenya (TRNPR) are included for comparison. Each point represents the fecal microbiome of an individual primate. Two-dimensional stress value indicates moderate fidelity of original Bray–Curtis similarity matrix as represented in 2D space.
Fig. 2
Non-metric multi-dimensional scaling ordination based on Bray–Curtis similarity matrix comparing the fecal microbiomes of people, livestock, and non-human primates (BW, black-and-white colobus; RC, red colobus; RT, red-tailed guenon) from the Kibale area. Red colobus from Tana River National Park, Kenya (TRNPR) are included for comparison. Each point represents the fecal microbiome of an individual. Two-dimensional stress value indicates moderate fidelity of original Bray–Curtis similarity matrix as represented in 2D space.
Similar articles
- Giardia sp. and Cryptosporidium sp. infections in primates in fragmented and undisturbed forest in western Uganda.
Salzer JS, Rwego IB, Goldberg TL, Kuhlenschmidt MS, Gillespie TR. Salzer JS, et al. J Parasitol. 2007 Apr;93(2):439-40. doi: 10.1645/GE-970R1.1. J Parasitol. 2007. PMID: 17539436 - Life on the edge: gastrointestinal parasites from the forest edge and interior primate groups.
Chapman CA, Speirs ML, Gillespie TR, Holland T, Austad KM. Chapman CA, et al. Am J Primatol. 2006 Apr;68(4):397-409. doi: 10.1002/ajp.20233. Am J Primatol. 2006. PMID: 16534810 - Characterization of the fecal microbiome from non-human wild primates reveals species specific microbial communities.
Yildirim S, Yeoman CJ, Sipos M, Torralba M, Wilson BA, Goldberg TL, Stumpf RM, Leigh SR, White BA, Nelson KE. Yildirim S, et al. PLoS One. 2010 Nov 12;5(11):e13963. doi: 10.1371/journal.pone.0013963. PLoS One. 2010. PMID: 21103066 Free PMC article. - Forests without primates: primate/plant codependency.
Chapman CA, Onderdonk DA. Chapman CA, et al. Am J Primatol. 1998;45(1):127-41. doi: 10.1002/(SICI)1098-2345(1998)45:1<127::AID-AJP9>3.0.CO;2-Y. Am J Primatol. 1998. PMID: 9573446 Review. - Predicting primate local extinctions within "real-world" forest fragments: a pan-neotropical analysis.
Benchimol M, Peres CA. Benchimol M, et al. Am J Primatol. 2014 Mar;76(3):289-302. doi: 10.1002/ajp.22233. Epub 2013 Nov 8. Am J Primatol. 2014. PMID: 24532182 Review.
Cited by
- The Evolution of Stomach Acidity and Its Relevance to the Human Microbiome.
Beasley DE, Koltz AM, Lambert JE, Fierer N, Dunn RR. Beasley DE, et al. PLoS One. 2015 Jul 29;10(7):e0134116. doi: 10.1371/journal.pone.0134116. eCollection 2015. PLoS One. 2015. PMID: 26222383 Free PMC article. Review. - Effects of anthropogenic habitat disturbance and Giardia duodenalis infection on a sentinel species' gut bacteria.
Kuthyar S, Kowalewski MM, Roellig DM, Mallott EK, Zeng Y, Gillespie TR, Amato KR. Kuthyar S, et al. Ecol Evol. 2020 Dec 12;11(1):45-57. doi: 10.1002/ece3.6910. eCollection 2021 Jan. Ecol Evol. 2020. PMID: 33437414 Free PMC article. - Diet components associated with specific bacterial taxa shape overall gut community compositions in omnivorous African viverrids.
Storm MB, Arfaoui EMR, Simelane P, Denlinger J, Dias CA, da Conceição AG, Monadjem A, Bohmann K, Poulsen M, Bodawatta KH. Storm MB, et al. Ecol Evol. 2024 Jul 11;14(7):e11486. doi: 10.1002/ece3.11486. eCollection 2024 Jul. Ecol Evol. 2024. PMID: 39005885 Free PMC article. - Evaluation of fecal microbiomes associated with obesity in captive cynomolgus monkeys (Macaca fascicularis).
Koo BS, Hwang EH, Kim G, Oh H, Son Y, Lee D, Lim KS, Kang P, Lee S, Lee HY, Jeong KJ, Lee Y, Baek SH, Jeon CY, Park SJ, Kim YH, Huh JW, Jin YB, Kim SU, Lee SR, Hong JJ. Koo BS, et al. J Vet Sci. 2019 May;20(3):e19. doi: 10.4142/jvs.2019.20.e19. J Vet Sci. 2019. PMID: 31161737 Free PMC article. - The critical role of dietary foliage in maintaining the gut microbiome and metabolome of folivorous sifakas.
Greene LK, McKenney EA, O'Connell TM, Drea CM. Greene LK, et al. Sci Rep. 2018 Sep 27;8(1):14482. doi: 10.1038/s41598-018-32759-7. Sci Rep. 2018. PMID: 30262842 Free PMC article.
References
- Banks JC, Cary SC, Hogg ID. The phylogeography of Adelie penguin faecal flora. Environ Microbiol. 2009;11:577–588. - PubMed
- Bonnell TR, Sengupta RR, Chapman CA, Goldberg TL. An agent-based model of red colobus resources and disease dynamics implicates key resource sites as hot spots of disease transmission. Ecol Model. 2010;221:2491–2500.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical