Genome-wide nucleosome map and cytosine methylation levels of an ancient human genome - PubMed (original) (raw)
doi: 10.1101/gr.163592.113. Epub 2013 Dec 3.
Eivind Valen, Amhed M Vargas Velazquez, Brian J Parker, Morten Rasmussen, Stinus Lindgreen, Berit Lilje, Desmond J Tobin, Theresa K Kelly, Søren Vang, Robin Andersson, Peter A Jones, Cindi A Hoover, Alexei Tikhonov, Egor Prokhortchouk, Edward M Rubin, Albin Sandelin, M Thomas P Gilbert, Anders Krogh, Eske Willerslev, Ludovic Orlando
Affiliations
- PMID: 24299735
- PMCID: PMC3941110
- DOI: 10.1101/gr.163592.113
Genome-wide nucleosome map and cytosine methylation levels of an ancient human genome
Jakob Skou Pedersen et al. Genome Res. 2014 Mar.
Abstract
Epigenetic information is available from contemporary organisms, but is difficult to track back in evolutionary time. Here, we show that genome-wide epigenetic information can be gathered directly from next-generation sequence reads of DNA isolated from ancient remains. Using the genome sequence data generated from hair shafts of a 4000-yr-old Paleo-Eskimo belonging to the Saqqaq culture, we generate the first ancient nucleosome map coupled with a genome-wide survey of cytosine methylation levels. The validity of both nucleosome map and methylation levels were confirmed by the recovery of the expected signals at promoter regions, exon/intron boundaries, and CTCF sites. The top-scoring nucleosome calls revealed distinct DNA positioning biases, attesting to nucleotide-level accuracy. The ancient methylation levels exhibited high conservation over time, clustering closely with modern hair tissues. Using ancient methylation information, we estimated the age at death of the Saqqaq individual and illustrate how epigenetic information can be used to infer ancient gene expression. Similar epigenetic signatures were found in other fossil material, such as 110,000- to 130,000-yr-old bones, supporting the contention that ancient epigenomic information can be reconstructed from a deep past. Our findings lay the foundation for extracting epigenomic information from ancient samples, allowing shifts in epialleles to be tracked through evolutionary time, as well as providing an original window into modern epigenomics.
Figures
Figure 1.
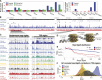
Paleo-Eskimo read depth reflects nucleosome occupancy. (A, left) Regional variation in read depth relative to genomic average (enrichment) for Saqqaq, Control, Aboriginal, and an experimental occupancy map (‘Schones’) (Schones et al. 2008). (Right) Saqqaq and Control regional read-depth variation after GC-correction. (B) Read-depth variation in a centromeric region known to harbor a 200-kb array of well-positioned nucleosomes (Gaffney et al. 2012) (left) and a region with genes (right). CpG islands (green bars) correlate with elevated read depth in the Saqqaq. The variation is also observed in genomically unique regions (black bars), where reads down to length 25 can map. The read depth of the Control exhibits lower variance. (C) Examples of Saqqaq read-depth variation, GC corrected read-depth variation, Saqqaq nucleosome predictions, and experimental (Schones, from CD4+ cells) as well as computational (Dennis and A375) (Dennis et al. 2007; Ozsolak et al. 2007) occupancy maps in ∼2-kb regions of the nucleosome array (left) and a transcription start site (TSS) region (right). Light gray denotes the 147-bp-long nucleosome predictions. Saqqaq read depth correlates with both the read depth of the ancient Aboriginal genome and the occupancy maps, but not with the Control. (D) DNA packaged around nucleosomes. We hypothesize that DNA wound around nucleosomes to be better protected from degradation. (E) The Saqqaq shows more variation in read depth than Control, with more genomic sites showing extremely low or high read depth. (F) Distribution of correlations for Saqqaq versus other sets across all promoter regions.
Figure 2.
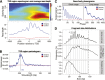
Read depth and fragment length periodicity. (A) Read-depth variation at TSS. Spectrogram around TSS (top) showing the strength of the periodicity signal at different wavelengths. Nucleosome abundance (bottom) summed over aligned transcription start sites. High occupancy at the +1 nucleosome position is characteristic of transcriptional activity. (B) Spectral density (periodogram) for TSS regions. The frequency spectrum shows a peak in relative signal at 193 bp corresponding to the expected inter-nucleosome distance. (C) 5′ read-end phasograms showing the distribution of distances between reads in gene bodies. A clear ∼200-bp periodicity is apparent, consistent with the presence of nucleosomes (right). A short-range periodicity of ∼10 bp is also apparent (left), corresponding to a turn of the DNA helix as it winds around the nucleosome. (D) Distribution of fragment sizes from ancient samples of horse (top), polar bear (middle), and Saqqaq (bottom) are consistent with preferential cleavage of exposed nucleosome-wrapped DNA strands every 10 bp.
Figure 3.
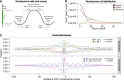
Nucleosome calls and positioning patterns. (A) Nucleosome center positions (dyads) are called as read-depth peaks if maximal at the center of a running window of nucleosome length 147 bp. Calls are scored by the difference in read depth between the peak (p) and the average read depth of the left (lf) and right (rf) flanking regions [score = p − (lf + rf)/2]. (B) Nucleosome call abundance is shown as a function of quality score cutoff for the Saqqaq (blue) and the Control (red), which lacks the nucleosome signal. The difference (green) gives the expected number of true positive calls at a given score cutoff and, indirectly, the FDR (<1% for the 1.9M calls with a score cutoff >29). (C) Base composition and distribution of purine/pyrimidine sequence dimers across the top 25% of called nucleosomes.
Figure 4.
Substitution rates at CpG reveal methylation of DNA. (A) C→T mismatch rates (gray) versus rate of other mismatches (black) between a random subset of 1,000,000 Phusion (left) or HiFi (right) reads mapping uniquely. Reads are split by those starting with CpG (top; 26,864 Phusion and 25,568 HiFi reads) and other dinucleotides (bottom). (B) Mismatch frequencies for Phusion (left) and HiFi (right) for reads aligned to various genomic locations starting with the dinucleotides: CpG (top) and Cp[ACT] (bottom). (C) Distribution of Ms values for three classes of promoters with low, medium, and high CpG densities (Supplemental Material SI3.2). (D) Methylation profile (Ms, top) and read depth variation (bottom) at CTCF regions. Read depth provides a proxy for nucleosome occupancy. (E) Distribution of Ms values across nucleotide positions covered with nucleosomes, showing a depletion in methylation levels within a core region (20 nt before and after the nucleosome center) that is particularly marked at the nucleosome center.
Figure 5.
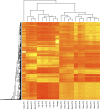
Unsupervised hierarchical clustering of tissue methylation profiles. Ms-based methylation levels of the Saqqaq individual are compared to the methylation profiles of five modern donors (PT1, PT2, PT3, PT4, and PT5) across four tissues (blood, buccal, saliva, and hair). Ms calculations were based on 2000-bp-wide genomic regions centered on each locus from the Illumina 450k array, disregarding those that showed less than 100 CpG sites at read starts (Supplemental Material SI3.4). The final set includes a total number of 7383 CpG sites.
Figure 6.
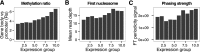
Nucleosome and methylation maps as proxies for ancient gene expression. Relationship between three measures assessing gene expression. (A) Methylation ratio (Rs), a measure of methylation in promoter versus gene bodies. (B) First nucleosome occupancy, average read depth over the TSS +1 nucleosome region. (C) Phasing strength, a measure of strength of the periodicity between neighboring nucleosomes across the TSS region by Fourier transform analysis. All display a significant correlation with expression as measured by microarrays in modern hair follicles.
Similar articles
- Fast, Accurate and Automatic Ancient Nucleosome and Methylation Maps with epiPALEOMIX.
Hanghøj K, Seguin-Orlando A, Schubert M, Madsen T, Pedersen JS, Willerslev E, Orlando L. Hanghøj K, et al. Mol Biol Evol. 2016 Dec;33(12):3284-3298. doi: 10.1093/molbev/msw184. Epub 2016 Sep 13. Mol Biol Evol. 2016. PMID: 27624717 Free PMC article. - Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules.
Kelly TK, Liu Y, Lay FD, Liang G, Berman BP, Jones PA. Kelly TK, et al. Genome Res. 2012 Dec;22(12):2497-506. doi: 10.1101/gr.143008.112. Epub 2012 Sep 7. Genome Res. 2012. PMID: 22960375 Free PMC article. - Understanding the connection between epigenetic DNA methylation and nucleosome positioning from computer simulations.
Portella G, Battistini F, Orozco M. Portella G, et al. PLoS Comput Biol. 2013;9(11):e1003354. doi: 10.1371/journal.pcbi.1003354. Epub 2013 Nov 21. PLoS Comput Biol. 2013. PMID: 24278005 Free PMC article. - Epigenomic Modifications in Modern and Ancient Genomes.
Niiranen L, Leciej D, Edlund H, Bernhardsson C, Fraser M, Quinto FS, Herzig KH, Jakobsson M, Walkowiak J, Thalmann O. Niiranen L, et al. Genes (Basel). 2022 Jan 20;13(2):178. doi: 10.3390/genes13020178. Genes (Basel). 2022. PMID: 35205223 Free PMC article. Review. - Studying the epigenome using next generation sequencing.
Ku CS, Naidoo N, Wu M, Soong R. Ku CS, et al. J Med Genet. 2011 Nov;48(11):721-30. doi: 10.1136/jmedgenet-2011-100242. Epub 2011 Aug 8. J Med Genet. 2011. PMID: 21825079 Review.
Cited by
- DNA methylation-based profiling of horse archaeological remains for age-at-death and castration.
Liu X, Seguin-Orlando A, Chauvey L, Tressières G, Schiavinato S, Tonasso-Calvière L, Aury JM, Perdereau A, Wagner S, Clavel P, Estrada O, Pan J, Ma Y, Enk J, Devault A, Klunk J, Lepetz S, Clavel B, Jiang L, Wincker P, Collin YRH, Sarkissian C, Orlando L. Liu X, et al. iScience. 2023 Feb 5;26(3):106144. doi: 10.1016/j.isci.2023.106144. eCollection 2023 Mar 17. iScience. 2023. PMID: 36843848 Free PMC article. - Recently evolved human-specific methylated regions are enriched in schizophrenia signals.
Banerjee N, Polushina T, Bettella F, Giddaluru S, Steen VM, Andreassen OA, Le Hellard S. Banerjee N, et al. BMC Evol Biol. 2018 May 11;18(1):63. doi: 10.1186/s12862-018-1177-2. BMC Evol Biol. 2018. PMID: 29747567 Free PMC article. - Novel Substrates as Sources of Ancient DNA: Prospects and Hurdles.
Green EJ, Speller CF. Green EJ, et al. Genes (Basel). 2017 Jul 13;8(7):180. doi: 10.3390/genes8070180. Genes (Basel). 2017. PMID: 28703741 Free PMC article. Review. - Prehistoric genomes reveal the genetic foundation and cost of horse domestication.
Schubert M, Jónsson H, Chang D, Der Sarkissian C, Ermini L, Ginolhac A, Albrechtsen A, Dupanloup I, Foucal A, Petersen B, Fumagalli M, Raghavan M, Seguin-Orlando A, Korneliussen TS, Velazquez AM, Stenderup J, Hoover CA, Rubin CJ, Alfarhan AH, Alquraishi SA, Al-Rasheid KA, MacHugh DE, Kalbfleisch T, MacLeod JN, Rubin EM, Sicheritz-Ponten T, Andersson L, Hofreiter M, Marques-Bonet T, Gilbert MT, Nielsen R, Excoffier L, Willerslev E, Shapiro B, Orlando L. Schubert M, et al. Proc Natl Acad Sci U S A. 2014 Dec 30;111(52):E5661-9. doi: 10.1073/pnas.1416991111. Epub 2014 Dec 15. Proc Natl Acad Sci U S A. 2014. PMID: 25512547 Free PMC article. - Epigenetic biomarkers and preterm birth.
Park B, Khanam R, Vinayachandran V, Baqui AH, London SJ, Biswal S. Park B, et al. Environ Epigenet. 2020 Jun 14;6(1):dvaa005. doi: 10.1093/eep/dvaa005. eCollection 2020 Jan. Environ Epigenet. 2020. PMID: 32551139 Free PMC article. Review.
References
- Bell AC, West AG, Felsenfeld G 2001. Insulators and boundaries: Versatile regulatory elements in the eukaryotic genome. Science 291: 447–450 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous