Virus Variation Resource--recent updates and future directions - PubMed (original) (raw)
. 2014 Jan;42(Database issue):D660-5.
doi: 10.1093/nar/gkt1268. Epub 2013 Dec 4.
Affiliations
- PMID: 24304891
- PMCID: PMC3965055
- DOI: 10.1093/nar/gkt1268
Virus Variation Resource--recent updates and future directions
J Rodney Brister et al. Nucleic Acids Res. 2014 Jan.
Abstract
Virus Variation (http://www.ncbi.nlm.nih.gov/genomes/VirusVariation/) is a comprehensive, web-based resource designed to support the retrieval and display of large virus sequence datasets. The resource includes a value added database, a specialized search interface and a suite of sequence data displays. Virus-specific sequence annotation and database loading pipelines produce consistent protein and gene annotation and capture sequence descriptors from sequence records then map these metadata to a controlled vocabulary. The database supports a metadata driven, web-based search interface where sequences can be selected using a variety of biological and clinical criteria. Retrieved sequences can then be downloaded in a variety of formats or analyzed using a suite of tools and displays. Over the past 2 years, the pre-existing influenza and Dengue virus resources have been combined into a single construct and West Nile virus added to the resultant resource. A number of improvements were incorporated into the sequence annotation and database loading pipelines, and the virus-specific search interfaces were updated to support more advanced functions. Several new features have also been added to the sequence download options, and a new multiple sequence alignment viewer has been incorporated into the resource tool set. Together these enhancements should support enhanced usability and the inclusion of new viruses in the future.
Figures
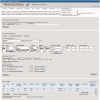
Figure 1.
Influenza virus database search interface. The search interface is shown with the ‘Additional filters’ selection open. A number of search criteria have been selected, and two separate searches added to the ‘Query builder’. Selected search results are indicated by the check box to the left of the ‘Query builder’ display and can be downloaded directly in a variety of formats or loaded into the Virus Variation search results interface by depressing the ‘Show results’ button.
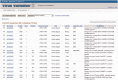
Figure 2.
Virus Variation search results interface. Results from a DENV database query are shown. The display includes a number of retrieved sequence descriptors including Accession, Length, Type, Disease, Genome region, Host, Country, Collection date and Virus name. Sequences can be selected and downloaded in a number of formats or used to construct an alignment or phylogenetic tree.
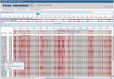
Figure 3.
Virus Variation multi-sequence alignment viewer. The results from a DENV database query were aligned and displayed in the Virus Variation multi-sequence alignment viewer. The top section of the alignment viewer includes a histogram that displays sequence and coverage variation across the alignment, a second histogram that plots the frequency of sequence differences with a shading scheme and highlights insertions and deletions with gaps and a feature table where protein names and other sequence feature identifiers are displayed. The alignment position is indicated above the histogram, and the region displayed in the lower section is highlighted by a gray box. The lower section displays the highlighted region in greater detail by default, but the magnification can be decreased or increased as desired by the user. Alignments are anchored to the consensus sequence by default, but any sequence can be selected as an anchor. Sequences identical to the consensus can be displayed as individual nucleotides or amino acids or replaced with dots—highlighting variations from the consensus.
Similar articles
- Virus Variation Resource - improved response to emergent viral outbreaks.
Hatcher EL, Zhdanov SA, Bao Y, Blinkova O, Nawrocki EP, Ostapchuck Y, Schäffer AA, Brister JR. Hatcher EL, et al. Nucleic Acids Res. 2017 Jan 4;45(D1):D482-D490. doi: 10.1093/nar/gkw1065. Epub 2016 Nov 28. Nucleic Acids Res. 2017. PMID: 27899678 Free PMC article. - Virus variation resources at the National Center for Biotechnology Information: dengue virus.
Resch W, Zaslavsky L, Kiryutin B, Rozanov M, Bao Y, Tatusova TA. Resch W, et al. BMC Microbiol. 2009 Apr 2;9:65. doi: 10.1186/1471-2180-9-65. BMC Microbiol. 2009. PMID: 19341451 Free PMC article. - Influenza Virus Database (IVDB): an integrated information resource and analysis platform for influenza virus research.
Chang S, Zhang J, Liao X, Zhu X, Wang D, Zhu J, Feng T, Zhu B, Gao GF, Wang J, Yang H, Yu J, Wang J. Chang S, et al. Nucleic Acids Res. 2007 Jan;35(Database issue):D376-80. doi: 10.1093/nar/gkl779. Epub 2006 Oct 25. Nucleic Acids Res. 2007. PMID: 17065465 Free PMC article. - NCBI Taxonomy: a comprehensive update on curation, resources and tools.
Schoch CL, Ciufo S, Domrachev M, Hotton CL, Kannan S, Khovanskaya R, Leipe D, Mcveigh R, O'Neill K, Robbertse B, Sharma S, Soussov V, Sullivan JP, Sun L, Turner S, Karsch-Mizrachi I. Schoch CL, et al. Database (Oxford). 2020 Jan 1;2020:baaa062. doi: 10.1093/database/baaa062. Database (Oxford). 2020. PMID: 32761142 Free PMC article. Review. - Navigating the Landscape: A Comprehensive Review of Current Virus Databases.
Ritsch M, Cassman NA, Saghaei S, Marz M. Ritsch M, et al. Viruses. 2023 Aug 29;15(9):1834. doi: 10.3390/v15091834. Viruses. 2023. PMID: 37766241 Free PMC article. Review.
Cited by
- Quantification of dengue virus specific T cell responses and correlation with viral load and clinical disease severity in acute dengue infection.
Wijeratne DT, Fernando S, Gomes L, Jeewandara C, Ginneliya A, Samarasekara S, Wijewickrama A, Hardman CS, Ogg GS, Malavige GN. Wijeratne DT, et al. PLoS Negl Trop Dis. 2018 Oct 1;12(10):e0006540. doi: 10.1371/journal.pntd.0006540. eCollection 2018 Oct. PLoS Negl Trop Dis. 2018. PMID: 30273352 Free PMC article. - Database resources of the National Center for Biotechnology Information.
NCBI Resource Coordinators. NCBI Resource Coordinators. Nucleic Acids Res. 2015 Jan;43(Database issue):D6-17. doi: 10.1093/nar/gku1130. Epub 2014 Nov 14. Nucleic Acids Res. 2015. PMID: 25398906 Free PMC article. - Functional Characterization of Adaptive Mutations during the West African Ebola Virus Outbreak.
Dietzel E, Schudt G, Krähling V, Matrosovich M, Becker S. Dietzel E, et al. J Virol. 2017 Jan 3;91(2):e01913-16. doi: 10.1128/JVI.01913-16. Print 2017 Jan 15. J Virol. 2017. PMID: 27847361 Free PMC article. - A single mutation in the envelope protein modulates flavivirus antigenicity, stability, and pathogenesis.
Goo L, VanBlargan LA, Dowd KA, Diamond MS, Pierson TC. Goo L, et al. PLoS Pathog. 2017 Feb 16;13(2):e1006178. doi: 10.1371/journal.ppat.1006178. eCollection 2017 Feb. PLoS Pathog. 2017. PMID: 28207910 Free PMC article. - Time-series oligonucleotide count to assign antiviral siRNAs with long utility fit in the big data era.
Wada K, Wada Y, Iwasaki Y, Ikemura T. Wada K, et al. Gene Ther. 2017 Oct;24(10):668-673. doi: 10.1038/gt.2017.76. Epub 2017 Sep 14. Gene Ther. 2017. PMID: 28905886 Free PMC article.
References
- Fauci AS. Race against time. Nature. 2005;435:423–424. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous