HMGA2 functions as a competing endogenous RNA to promote lung cancer progression - PubMed (original) (raw)
HMGA2 functions as a competing endogenous RNA to promote lung cancer progression
Madhu S Kumar et al. Nature. 2014.
Retraction in
- Retraction: HMGA2 functions as a competing endogenous RNA to promote lung cancer progression.
Kumar MS, Armenteros-Monterroso E, East P, Chakravorty P, Matthews N, Winslow MM, Downward J. Kumar MS, et al. Nature. 2015 Jul 16;523(7560):370. doi: 10.1038/nature14551. Epub 2015 Jun 10. Nature. 2015. PMID: 26061768 Free PMC article. No abstract available.
Abstract
Non-small-cell lung cancer (NSCLC) is the most prevalent histological cancer subtype worldwide. As the majority of patients present with invasive, metastatic disease, it is vital to understand the basis for lung cancer progression. Hmga2 is highly expressed in metastatic lung adenocarcinoma, in which it contributes to cancer progression and metastasis. Here we show that Hmga2 promotes lung cancer progression in mouse and human cells by operating as a competing endogenous RNA (ceRNA) for the let-7 microRNA (miRNA) family. Hmga2 can promote the transformation of lung cancer cells independent of protein-coding function but dependent upon the presence of let-7 sites; this occurs without changes in the levels of let-7 isoforms, suggesting that Hmga2 affects let-7 activity by altering miRNA targeting. These effects are also observed in vivo, where Hmga2 ceRNA activity drives lung cancer growth, invasion and dissemination. Integrated analysis of miRNA target prediction algorithms and metastatic lung cancer gene expression data reveals the TGF-β co-receptor Tgfbr3 (ref. 12) as a putative target of Hmga2 ceRNA function. Tgfbr3 expression is regulated by the Hmga2 ceRNA through differential recruitment to Argonaute 2 (Ago2), and TGF-β signalling driven by Tgfbr3 is important for Hmga2 to promote lung cancer progression. Finally, analysis of NSCLC-patient gene-expression data reveals that HMGA2 and TGFBR3 are coordinately regulated in NSCLC-patient material, a vital corollary to ceRNA function. Taken together, these results suggest that Hmga2 promotes lung carcinogenesis both as a protein-coding gene and as a non-coding RNA; such dual-function regulation of gene-expression networks reflects a novel means by which oncogenes promote disease progression.
Figures
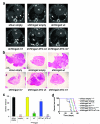
Figure 1. Hmga2 promotes lung cancer cell transformation in a protein-coding independent but let-7 site dependent manner
a, Diagram of Hmga2 allelic series: expression constructs containing the entire Hmga2 cDNA (“wt”); the cDNA with all seven let-7 sites in the 3′ UTR mutated (“m7”); the cDNA with the start codon mutated (“ATG wt”); and the cDNA with both the start codon and let-7 sites mutated (“ATG m7”). b, Hmga2 wt and m7 induce Hmga2 expression in non-metastatic lung cancer cells (368T1) and restores expression cells in metastatic lung cancer cells (482N1) depleted for endogenous Hmga2 (shHmga2). Two distinct HMGA2 antibodies are used: one recognizes the N-terminus of HMGA2 (HMGA2-CST) and the other recognizes the central AT-hook region of HMGA2 (HMGA2-Narita). c, Hmga2 RNA is comparably expressed by the wt, m7, ATG wt, and ATG m7 in both 368T1 and 482N1 cells. Hmga2 expression is normalized to Gapdh. 368T1 values are normalized to empty and 482N1 values are normalized to shluc empty. Values are technical triplicates, have been performed independently three times, and represent mean +/− standard deviation (s.d.) with propagated error. d, Hmga2 wt and ATG wt promote substantial anchorage-independent growth in both 368T1 and 482N1 cells. Values are technical triplicates, have been performed independently three times, and represent mean +/− s.d. e, Representative images of soft agar colonies. Magnification is 10X. ***: p<0.0005; **: p<0.005; *: p<0.05.
Figure 2. Hmga2 ceRNA activity enhances lung cancer progression in vivo
a, Hmga2 wt and ATG wt restore lung tumour growth in response to endogenous Hmga2 knockdown. B6129SF1/Tac males were intravenously injected with 482N1 cells expressing either a control shRNA and empty vector (shluc empty) or shHmga2 with the Hmga2 allelic series. Three weeks afterwards, animals were scanned by micro-CT and representative transverse images are shown. The heart is demarcated (labelled ‘H’) and white arrows identify lung tumours. b, Representative histological images of lungs transplanted with 482N1 cells from the series described in a. Magnification is 1X. c, Lung surface tumour counts were taken from animals transplanted with 482N1 cells from the series described in a (n=3 animals per group). Values are technical triplicates represent mean +/− s.e.m. d, Hmga2 wt and ATG wt substantially reduce survival of animals transplanted with 482N1 cells expressing the shRNA targeting Hmga2. Animals were intravenously transplanted with cells as in a. Animals were subsequently aged for survival and a Kaplan-Meier analysis was performed (n=9 animals per group). Median survival was 34 days for shluc empty/shHmga2 wt transplants; 37 days for shHmga2 ATG wt transplants; 43 days for shHmga2 m7 transplants; and 50 days for shHmga2 empty/ATG m7 transplants. Statistical significance was assessed by log-rank tests compared to shHmga2 empty. ****: p<0.00005; ***: p<0.0005; **: p<0.005; *: p<0.05; n.s.: not significant.
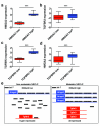
Figure 3. Hmga2 ceRNA activity enhances TGF-β signalling through over-expression of Tgfbr3
a, Hmga2 wt and ATG wt substantially induce both Tgfbr3 protein expression and phosphorylation of Smad2 (pSmad2) in both 368T1 and 482N1 cells. b, Hmga2 wt and ATG wt significantly promote expression of Tgfbr3 mRNA in both 368T1 and 482N1 cells. Tgfbr3 expression is normalized to Gapdh. 368T1 values are normalized to empty and 482N1 values are normalized to shluc empty. Values are technical triplicates, have been performed independently three times, and represent mean +/− s.d. with propagated error. c, Hmga2 wt and ATG wt specifically induce expression of a luciferase Tgfbr3 3′ UTR reporter in a let-7 site-dependent manner in both 368T1 and 482N1 cells. Cells were transfected with Renilla constructs of the control siCXCR4 multimer and either the Tgfbr3 wild type or let-7 mutant 3′ UTR reporter. Values are normalized to co-transfected pGL3 plasmid. 368T1 values are normalized to empty and 482N1 values are normalized to shluc empty. Values are technical triplicates, have been performed independently three times, and represent mean +/− s.d. with propagated error. d, Hmga2 wt and ATG wt displace Tgfbr3 from Argonaute-2 (Ago2) based RNA-induced silencing complexes. Lysates from 368T1 and 482N1 cells of the Hmga2 allelic series underwent either control immunoprecipitation (IgG) or immunoprecipitation for Ago2. RNA was purified and qRT-PCR was performed for Hmga2 and Tgfbr3 on both the immunoprecipitated and input RNAs. Values are depicted as the percentage of input RNA, are technical triplicates, have been performed independently twice, and represent mean +/− s.d. e, multiple shRNAs elicit substantial knockdown of Tgfbr3 mRNA in both 368T1 and 482N1 cells. 482N1 cells were infected with control shRNA (shluc) or a set of shRNAs targeting Tgfbr3 (shTgfbr3.1-3.5), while 368T1 wt and ATG wt cells were infected with shluc or shTgfbr3.1, 3.2, 3.4, and 3.5. RNA was purified and qRT-PCR was performed. Tgfbr3 expression is normalized to Gapdh and 368T1 wt and ATG wt and 482N1 values are normalized to shluc. Values are technical triplicates, have been performed independently three times, and represent mean +/− s.d. with propagated error. f, multiple shRNAs induce knockdown of Tgbr3 and suppress TGF-β pathway activity in 368T1 and 482N1 cells. Cells were infected with shRNAs as in e and Western analysis was performed for Tgfbr3, pSmad2 and total Smad2 (Smad2). g, Tgfbr3 depletion reduces anchorage-independent growth of 368T1 wt and ATG wt and 482N1 cells. Cells were infected with the listed shRNAs and plated for anchorage-independent growth and colonies were counted as above. Values are technical triplicates, have been performed independently three times, and represent mean +/− s.d. ***: p<0.0005; **: p<0.005; *: p<0.05.
Figure 4. HMGA2 and TGFBR3 are reciprocally and co-ordinately upregulated in NSCLC patients
a, The Cancer Genome Atlas (TCGA) NSCLC gene expression data set was sorted on HMGA2 expression. The top and bottom quartiles (HMGA2 low and high, respectively) were selected (45 patients per group) and HMGA2 expression was compared by box and whisker plot. b, The TCGA data set was sorted into top and bottom quartiles of HMGA2 expression as in a, and TGFBR3 expression was compared by box and whisker plot. c, The TCGA data set was sorted into top and bottom quartiles of TGFBR3 expression (TGFBR3 low and high, respectively) as in a, and TGFBR3 expression was compared by box and whisker plot. d, The TCGA data set was sorted into top and bottom quartiles of TGFBR3 expression as in c, and HMGA2 expression was compared by box and whisker plot. In all box and whisker plots, values are presented on a log2 scale. Significance was assessed by the Mann-Whitney test with a Bonferroni correction for multiple hypothesis testing. ***: p<0.0005. e, Model for Hmga2 acting as a competing endogenous RNA for Tgfbr3. In nonmetastatic NSCLC, Hmga2 expression is low, leading to suppressed Tgfbr3 expression by let-7. In metastatic NSCLC, Hmga2 expression is elevated, titrating away let-7 from Tgfbr3 and allowing for its over-expression. This titration occurs without changes in let-7 expression, reflecting competition for microRNA occupancy by target transcripts.
Comment in
- Lung cancer: A surprising competitor.
McCarthy N. McCarthy N. Nat Rev Cancer. 2014 Feb;14(2):73. doi: 10.1038/nrc3676. Nat Rev Cancer. 2014. PMID: 24457413 No abstract available. - Aberrant ceRNA activity drives lung cancer.
Tay Y, Karreth FA, Pandolfi PP. Tay Y, et al. Cell Res. 2014 Mar;24(3):259-60. doi: 10.1038/cr.2014.21. Epub 2014 Feb 14. Cell Res. 2014. PMID: 24525785 Free PMC article.
Similar articles
- MUC1-C Induces the LIN28B→LET-7→HMGA2 Axis to Regulate Self-Renewal in NSCLC.
Alam M, Ahmad R, Rajabi H, Kufe D. Alam M, et al. Mol Cancer Res. 2015 Mar;13(3):449-60. doi: 10.1158/1541-7786.MCR-14-0363. Epub 2014 Nov 3. Mol Cancer Res. 2015. PMID: 25368430 Free PMC article. - MicroRNA-219 is downregulated in non-small cell lung cancer and inhibits cell growth and metastasis by targeting HMGA2.
Sun X, Xu M, Liu H, Ming K. Sun X, et al. Mol Med Rep. 2017 Sep;16(3):3557-3564. doi: 10.3892/mmr.2017.7000. Epub 2017 Jul 15. Mol Med Rep. 2017. PMID: 28714014 Retracted. - The long non-coding RNA LSINCT5 promotes malignancy in non-small cell lung cancer by stabilizing HMGA2.
Tian Y, Zhang N, Chen S, Ma Y, Liu Y. Tian Y, et al. Cell Cycle. 2018;17(10):1188-1198. doi: 10.1080/15384101.2018.1467675. Epub 2018 Jul 5. Cell Cycle. 2018. PMID: 29883241 Free PMC article. - The Role of TGFBR3 in the Development of Lung Cancer.
Deng X, Ma N, He J, Xu F, Zou G. Deng X, et al. Protein Pept Lett. 2024;31(7):491-503. doi: 10.2174/0109298665315841240731060636. Protein Pept Lett. 2024. PMID: 39092729 Review. - MiRNAs and LncRNAs: Dual Roles in TGF-β Signaling-Regulated Metastasis in Lung Cancer.
Lai XN, Li J, Tang LB, Chen WT, Zhang L, Xiong LX. Lai XN, et al. Int J Mol Sci. 2020 Feb 11;21(4):1193. doi: 10.3390/ijms21041193. Int J Mol Sci. 2020. PMID: 32054031 Free PMC article. Review.
Cited by
- Cell states and neighborhoods in distinct clinical stages of primary and metastatic esophageal adenocarcinoma.
Yates J, Mathey-Andrews C, Park J, Garza A, Gagné A, Hoffman S, Bi K, Titchen B, Hennessey C, Remland J, Shannon E, Camp S, Balamurali S, Cavale SK, Li Z, Raghawan AK, Kraft A, Boland G, Aguirre AJ, Sethi NS, Boeva V, Van Allen E. Yates J, et al. bioRxiv [Preprint]. 2024 Aug 20:2024.08.17.608386. doi: 10.1101/2024.08.17.608386. bioRxiv. 2024. PMID: 39229240 Free PMC article. Preprint. - Dysregulation of TCONS_00006091 contributes to the elevated risk of oral squamous cell carcinoma by upregulating SNAI1, IRS and HMGA2.
Ma D, Chen J, Shi Y, Gao H, Wei Z, Fan J, Wang L. Ma D, et al. Sci Rep. 2024 Apr 26;14(1):9616. doi: 10.1038/s41598-024-60310-4. Sci Rep. 2024. PMID: 38671227 Free PMC article. - MicroRNAs in the Pathogenesis, Prognostication and Prediction of Treatment Resistance in Soft Tissue Sarcomas.
Teo AYT, Lim VY, Yang VS. Teo AYT, et al. Cancers (Basel). 2023 Jan 18;15(3):577. doi: 10.3390/cancers15030577. Cancers (Basel). 2023. PMID: 36765536 Free PMC article. Review. - Novel perspectives on antisense transcription in HIV-1, HTLV-1, and HTLV-2.
Lin E, Panfil AR, Sandel G, Jain P. Lin E, et al. Front Microbiol. 2022 Dec 23;13:1042761. doi: 10.3389/fmicb.2022.1042761. eCollection 2022. Front Microbiol. 2022. PMID: 36620051 Free PMC article. Review. - Systemic analysis identifying PVT1/DUSP13 axis for microvascular invasion in hepatocellular carcinoma.
Su R, Zhang H, Zhang L, Khan AR, Zhang X, Wang R, Shao C, Wei X, Xu X. Su R, et al. Cancer Med. 2023 Apr;12(7):8937-8955. doi: 10.1002/cam4.5546. Epub 2022 Dec 16. Cancer Med. 2023. PMID: 36524545 Free PMC article.
References
- Ferlay J, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. - PubMed
- Meyer B, et al. HMGA2 overexpression in non-small cell lung cancer. Mol Carcinog. 2007;46:503–511. - PubMed
- Sarhadi VK, et al. Increased expression of high mobility group A proteins in lung cancer. J Pathol. 2006;209:206–212. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 13-0142/AICR_/Worldwide Cancer Research/United Kingdom
- 323145/ERC_/European Research Council/International
- A3570/CRUK_/Cancer Research UK/United Kingdom
- A7419/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials