Genome-wide and paternal diversity reveal a recent origin of human populations in North Africa - PubMed (original) (raw)
Genome-wide and paternal diversity reveal a recent origin of human populations in North Africa
Karima Fadhlaoui-Zid et al. PLoS One. 2013.
Abstract
The geostrategic location of North Africa as a crossroad between three continents and as a stepping-stone outside Africa has evoked anthropological and genetic interest in this region. Numerous studies have described the genetic landscape of the human population in North Africa employing paternal, maternal, and biparental molecular markers. However, information from these markers which have different inheritance patterns has been mostly assessed independently, resulting in an incomplete description of the region. In this study, we analyze uniparental and genome-wide markers examining similarities or contrasts in the results and consequently provide a comprehensive description of the evolutionary history of North Africa populations. Our results show that both males and females in North Africa underwent a similar admixture history with slight differences in the proportions of admixture components. Consequently, genome-wide diversity show similar patterns with admixture tests suggesting North Africans are a mixture of ancestral populations related to current Africans and Eurasians with more affinity towards the out-of-Africa populations than to sub-Saharan Africans. We estimate from the paternal lineages that most North Africans emerged ∼15,000 years ago during the last glacial warming and that population splits started after the desiccation of the Sahara. Although most North Africans share a common admixture history, the Tunisian Berbers show long periods of genetic isolation and appear to have diverged from surrounding populations without subsequent mixture. On the other hand, continuous gene flow from the Middle East made Egyptians genetically closer to Eurasians than to other North Africans. We show that genetic diversity of today's North Africans mostly captures patterns from migrations post Last Glacial Maximum and therefore may be insufficient to inform on the initial population of the region during the Middle Paleolithic period.
Conflict of interest statement
Competing Interests: The authors have declare that no compiting interests exist.
Figures
Figure 1. Frequency of the major Y-chromosome haplogroups in North Africa and surrounding regions.
Intensity of the colors reflects the frequency of a haplogroup in the studied populations. A) Location of the analyzed populations. B–F) Frequency distribution of haplogroups E-M81, E-M78, E-M123, J-M267, and J-M172 respectively.
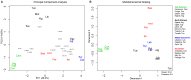
Figure 2. Y-chromosome population structure.
A) Principal component analysis of haplogroups frequencies. B) Multidimensional scaling plot based on RST distances between populations derived from Y-STR data.
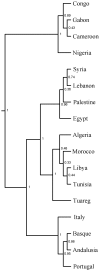
Figure 3. BATWING population splitting tree.
Numbers on branches show partition posterior probability.
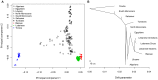
Figure 4. Genome-wide population structure.
A) Principal component analysis of ∼44,000 SNPs showing the top two components. B) Maximum likelihood tree showing populations relationships.
Similar articles
- Recent Historical Migrations Have Shaped the Gene Pool of Arabs and Berbers in North Africa.
Arauna LR, Mendoza-Revilla J, Mas-Sandoval A, Izaabel H, Bekada A, Benhamamouch S, Fadhlaoui-Zid K, Zalloua P, Hellenthal G, Comas D. Arauna LR, et al. Mol Biol Evol. 2017 Feb 1;34(2):318-329. doi: 10.1093/molbev/msw218. Mol Biol Evol. 2017. PMID: 27744413 Free PMC article. - Genome-wide diversity in the levant reveals recent structuring by culture.
Haber M, Gauguier D, Youhanna S, Patterson N, Moorjani P, Botigué LR, Platt DE, Matisoo-Smith E, Soria-Hernanz DF, Wells RS, Bertranpetit J, Tyler-Smith C, Comas D, Zalloua PA. Haber M, et al. PLoS Genet. 2013;9(2):e1003316. doi: 10.1371/journal.pgen.1003316. Epub 2013 Feb 28. PLoS Genet. 2013. PMID: 23468648 Free PMC article. - Genomic ancestry of North Africans supports back-to-Africa migrations.
Henn BM, Botigué LR, Gravel S, Wang W, Brisbin A, Byrnes JK, Fadhlaoui-Zid K, Zalloua PA, Moreno-Estrada A, Bertranpetit J, Bustamante CD, Comas D. Henn BM, et al. PLoS Genet. 2012 Jan;8(1):e1002397. doi: 10.1371/journal.pgen.1002397. Epub 2012 Jan 12. PLoS Genet. 2012. PMID: 22253600 Free PMC article. - African genetic diversity provides novel insights into evolutionary history and local adaptations.
Choudhury A, Aron S, Sengupta D, Hazelhurst S, Ramsay M. Choudhury A, et al. Hum Mol Genet. 2018 Aug 1;27(R2):R209-R218. doi: 10.1093/hmg/ddy161. Hum Mol Genet. 2018. PMID: 29741686 Free PMC article. Review. - Evolutionary Genetics and Admixture in African Populations.
Pfennig A, Petersen LN, Kachambwa P, Lachance J. Pfennig A, et al. Genome Biol Evol. 2023 Apr 6;15(4):evad054. doi: 10.1093/gbe/evad054. Genome Biol Evol. 2023. PMID: 36987563 Free PMC article. Review.
Cited by
- Human Dispersal Out of Africa: A Lasting Debate.
López S, van Dorp L, Hellenthal G. López S, et al. Evol Bioinform Online. 2016 Apr 21;11(Suppl 2):57-68. doi: 10.4137/EBO.S33489. eCollection 2015. Evol Bioinform Online. 2016. PMID: 27127403 Free PMC article. Review. - Carriers of Mitochondrial DNA Macrohaplogroup N Lineages Reached Australia around 50,000 Years Ago following a Northern Asian Route.
Fregel R, Cabrera V, Larruga JM, Abu-Amero KK, González AM. Fregel R, et al. PLoS One. 2015 Jun 8;10(6):e0129839. doi: 10.1371/journal.pone.0129839. eCollection 2015. PLoS One. 2015. PMID: 26053380 Free PMC article. - Novel Mutations Causing C5 Deficiency in Three North-African Families.
Colobran R, Franco-Jarava C, Martín-Nalda A, Baena N, Gabau E, Padilla N, de la Cruz X, Pujol-Borrell R, Comas D, Soler-Palacín P, Hernández-González M. Colobran R, et al. J Clin Immunol. 2016 May;36(4):388-96. doi: 10.1007/s10875-016-0275-4. Epub 2016 Mar 30. J Clin Immunol. 2016. PMID: 27026170 - Recent Historical Migrations Have Shaped the Gene Pool of Arabs and Berbers in North Africa.
Arauna LR, Mendoza-Revilla J, Mas-Sandoval A, Izaabel H, Bekada A, Benhamamouch S, Fadhlaoui-Zid K, Zalloua P, Hellenthal G, Comas D. Arauna LR, et al. Mol Biol Evol. 2017 Feb 1;34(2):318-329. doi: 10.1093/molbev/msw218. Mol Biol Evol. 2017. PMID: 27744413 Free PMC article. - The peopling of the last Green Sahara revealed by high-coverage resequencing of trans-Saharan patrilineages.
D'Atanasio E, Trombetta B, Bonito M, Finocchio A, Di Vito G, Seghizzi M, Romano R, Russo G, Paganotti GM, Watson E, Coppa A, Anagnostou P, Dugoujon JM, Moral P, Sellitto D, Novelletto A, Cruciani F. D'Atanasio E, et al. Genome Biol. 2018 Feb 12;19(1):20. doi: 10.1186/s13059-018-1393-5. Genome Biol. 2018. PMID: 29433568 Free PMC article.
References
- Barton RNE, Bouzouggar A, Collcutt SN, Schwenninger J-L, Clark-Balzan L (2009) OSL dating of the Aterian levels at Grotte de Dar es-Soltan I (Rabat, Morocco) and possible implications for the dispersal of modern Homo sapiens. Quaternary Sci Rev 28..
- Garcea EAA (2010) The spread of Aterian peoples in North Africa. In: GarceaEAA, editor. South-Eastern Mediterranean Peoples Between 130,000 and 10,000 years ago. Oxford: Oxbow Books.
- Debénath A (2000) Le peuplement préhistorique du Maroc: données récentes et problèmes. L'anthropologie 104: 131–145.
- Camps G (1974) Les civilisations préhistoriques de l'Afrique du Nord et du Sahara. Paris: Doin.
Publication types
MeSH terms
Grants and funding
This study was supported in parts by Spanish Government MCINN grant CGL2010-14944/BOS and Programa de Cooperación Interuniversitaria e Investigación Científica, Spanish Ministry of Foreign Affairs and Cooperation grants A75180/06, A/8394/07, B/018514/08, A1/040218/11. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources