Binding of the rhesus TRIM5α PRYSPRY domain to capsid is necessary but not sufficient for HIV-1 restriction - PubMed (original) (raw)
Binding of the rhesus TRIM5α PRYSPRY domain to capsid is necessary but not sufficient for HIV-1 restriction
Yang Yang et al. Virology. 2014.
Abstract
The PRYSPRY domain of TRIM5α provides specificity and the capsid recognition motif to retroviral restriction. Restriction of HIV-1 by rhesus TRIM5α (TRIM5αrh) has been correlated to its ability to bind to the HIV-1 core, suggesting that capsid binding is required for restriction. This work explores whether the PRYSPRY domain of TRIM5αrh exhibits an additional function besides binding to the HIV-1 core. Using our recently described structure of the PRYSPRY domain, we performed an exhaustive structure-function study of the surface and interior residues of the PRYSPRY domain. Testing retroviral restriction and capsid binding of an extensive collection of 60 TRIM5αrh PRYSPRY variants revealed that binding is necessary but not sufficient for restriction. In support of this hypothesis, we showed that some human tripartite motif proteins bind the HIV-1 capsid but do not restrict HIV-1 infection, such as human TRIM6 and TRIM34. Overall this work suggested that the PRYSPRY domain serves an unknown function, distinct from the binding of TRIM5αrh to the HIV-1 core, to block HIV-1 infection.
Keywords: Capsid; HIV-1; PRYSPRY; Restriction; TRIM5; V1 loop.
© 2013 Elsevier Inc. All rights reserved.
Figures
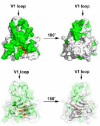
Figure 1. Structure of the PRYSPRY of TRIM5αrh showing the residues targeted in this study
The upper structures show the surface of the PRYSPRY domain of TRIM5αrh. Surface and internal residues targeted for mutagenesis are shown in green and orange, respectively. The lower structures show a transparent surface revealing the polypeptide chain of the TRIM5αrh PRYSPRY that similarly labels surface and internal residues targeted for mutagenesis in green and orange, respectively.
Figure 2. Expression of TRIM5αrh PRYSPRY domain variants
Dog Cf2Th cells were transduced with the LPCX vector expressing HA-tagged wild-type and mutant TRIM5αrh proteins. Stable cell lines were selected with 5 μg/ml of puromycin, and the expression levels of mutant and wild-type TRIM5α proteins were assayed by Western blotting using anti-HA antibodies. To control for protein loading, samples were Western blotted using anti-GAPDH antibodies. The results of three independent experiments were similar, and the results of a single experiment are shown.
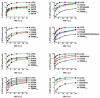
Figure 3. Restriction of HIV-1 infection by TRIM5αrh PRYSPRY variants
Dog Cf2Th cells stably expressing the different TRIM5αrh PRYSPRY variants were challenged with increasing amounts of HIV-1-GFP. Infection was determined by measuring the number of GFP-positive cells 48 hours post-infection using a flow cytometer. The graphs are organized containing mutants from potent to less potent restriction (top to button). Similar results were obtained in three independent experiments, and the result of one experiment is shown.
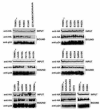
Figure 4. Binding of TRIM5αrh PRYSPRY variants to in vitro assembled HIV-1 capsid–nucleocapsid complexes
Human 293T cells were transfected with plasmids expressing the indicated wild-type and mutant HA-tagged TRIM5αrh proteins. Forty-eight hours after transfection, cells were lysed. The lysates were incubated at room temperature for 1 h together with in vitro assembled HIV-1 CA-NC complexes. The mixtures were applied to a 70% sucrose cushion and centrifuged. INPUT represents the lysates analyzed by Western blotting before being applied to the 70% cushion. The input mixtures were Western blotted using anti-HA antibodies. Similarly, the pellets from the 70% cushion, which represents the BOUND fraction**,** were analyzed by Western blotting using anti-HA and anti-p24 antibodies. The results of three independent experiments were similar; the result of a single experiment is shown.
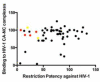
Figure 5. Binding of Trim5αrh to HIV-1 capsid is necessary but not sufficient for restriction
The relative binding of the different TRIM5αrh PRYSPRY domain variants to in vitro assembled HIV-1 CA-NC complexes was calculated as described in Table 1. Similarly, the ability of the different TRIM5αrh PRYSPRY domain variants to restrict HIV-1 was calculated as described in Table 1. The values for relative binding to HIV-1 CA-NC complexes were plotted as a function of HIV-1 restriction. Dots in red and yellow a represent TRIM5αrh PRYSPRY domain variants that are located on the surface patches SP1 and SP2, respectively.
Figure 6. TRIM5αrh PRYSPRY domain residues that are important for restriction but dispensable for binding to HIV-1 capsid
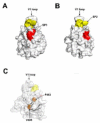
(A) The surface patch 1 (SP1), composed of D318, K319 and R320, is shown in red. (B) The surface patch 2 (SP2), composed of N326, P327, Q328, I329, M330 and Y331, is shown in yellow. (C) The polypeptide chain of the TRIM5αrh PRYSPRY domain showing the internal residues V405 and P483 in orange. In addition the structure shows SP1 and SP2 in red and yellow, respectively. (D) The conservation of SP1 and SP2 is shown in human and different species of primates such as rhesus macaques (rhesus m.), African green monkey pyg [agm (pyg)], African green monkey Tantalus [agm (tan)], orangutan, chimpanzee, spider monkey, tamarin monkey and squirrel monkey. The number for the rhesus macaque protein sequence is shown.
Figure 6. TRIM5αrh PRYSPRY domain residues that are important for restriction but dispensable for binding to HIV-1 capsid
(A) The surface patch 1 (SP1), composed of D318, K319 and R320, is shown in red. (B) The surface patch 2 (SP2), composed of N326, P327, Q328, I329, M330 and Y331, is shown in yellow. (C) The polypeptide chain of the TRIM5αrh PRYSPRY domain showing the internal residues V405 and P483 in orange. In addition the structure shows SP1 and SP2 in red and yellow, respectively. (D) The conservation of SP1 and SP2 is shown in human and different species of primates such as rhesus macaques (rhesus m.), African green monkey pyg [agm (pyg)], African green monkey Tantalus [agm (tan)], orangutan, chimpanzee, spider monkey, tamarin monkey and squirrel monkey. The number for the rhesus macaque protein sequence is shown.
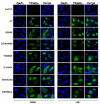
Figure 7. Intracellular distribution of TRIM5αrh PRYSPRY domain variants in the presence of leptomycin B
Human HeLa cells expressing the indicated TRIM5αrh variants were treated with 20 ng/ml of leptomycin B (LMB) or DMSO for 5 h. Treated cells were fixed and immunostained using anti-HA antibodies (green). Nuclei were stained with DAPI (blue). The results of three independent experiments were similar and a representative figure is shown. Quantification is shown in Figure S4.
Figure 8. Tripartite motif proteins that bind HIV-1 capsid but do not restrict HIV-1 infection
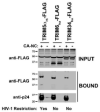
(A) The ability of TRIM6 and TRIM34 to bind in vitro assembled HIV-1 CA-NC complexes was performed, as described in Materials and Methods. Briefly, lysates containing TRIM6-FLAG or TRIM34-FLAG were incubated at room temperature for 1 h together with in vitro assembled HIV-1 CA-NC complexes. The mixtures were applied to a 70% sucrose cushion and centrifuged. INPUT represents the lysates analyzed by Western blotting before being applied to the 70% cushion. The input mixtures were Western blotted using anti-HA antibodies. Similarly, the pellets from the 70% cushion, which represents the BOUND fraction**,** were analyzed by Western blotting using anti-HA and anti-p24 antibodies. The results of three independent experiments were similar; the result of a single experiment is shown. (B) Similarly, we tested the ability of TRIM5α proteins from the indicated monkeys to bind in vitro assembled HIV-1 CA-NC complexes. The ability of these proteins to restrict HIV-1 is indicated in the bottom of the figure. The results of three independent experiments were similar and a representative figure is shown. The PRYSPRY domain of TRIM5αrh provides an unknown function to HIV-1 restriction. The PRYSPRY is important for the ability of TRIM5αrh to bind HIV-1 capsid. Human TRIM6 and TRIM34 bind HIV-1 capsid but don’t restrict HIV-1 infection.
Figure 8. Tripartite motif proteins that bind HIV-1 capsid but do not restrict HIV-1 infection
(A) The ability of TRIM6 and TRIM34 to bind in vitro assembled HIV-1 CA-NC complexes was performed, as described in Materials and Methods. Briefly, lysates containing TRIM6-FLAG or TRIM34-FLAG were incubated at room temperature for 1 h together with in vitro assembled HIV-1 CA-NC complexes. The mixtures were applied to a 70% sucrose cushion and centrifuged. INPUT represents the lysates analyzed by Western blotting before being applied to the 70% cushion. The input mixtures were Western blotted using anti-HA antibodies. Similarly, the pellets from the 70% cushion, which represents the BOUND fraction**,** were analyzed by Western blotting using anti-HA and anti-p24 antibodies. The results of three independent experiments were similar; the result of a single experiment is shown. (B) Similarly, we tested the ability of TRIM5α proteins from the indicated monkeys to bind in vitro assembled HIV-1 CA-NC complexes. The ability of these proteins to restrict HIV-1 is indicated in the bottom of the figure. The results of three independent experiments were similar and a representative figure is shown. The PRYSPRY domain of TRIM5αrh provides an unknown function to HIV-1 restriction. The PRYSPRY is important for the ability of TRIM5αrh to bind HIV-1 capsid. Human TRIM6 and TRIM34 bind HIV-1 capsid but don’t restrict HIV-1 infection.
Similar articles
- General Model for Retroviral Capsid Pattern Recognition by TRIM5 Proteins.
Wagner JM, Christensen DE, Bhattacharya A, Dawidziak DM, Roganowicz MD, Wan Y, Pumroy RA, Demeler B, Ivanov DN, Ganser-Pornillos BK, Sundquist WI, Pornillos O. Wagner JM, et al. J Virol. 2018 Jan 30;92(4):e01563-17. doi: 10.1128/JVI.01563-17. Print 2018 Feb 15. J Virol. 2018. PMID: 29187540 Free PMC article. - Structure of the rhesus monkey TRIM5α PRYSPRY domain, the HIV capsid recognition module.
Biris N, Yang Y, Taylor AB, Tomashevski A, Guo M, Hart PJ, Diaz-Griffero F, Ivanov DN. Biris N, et al. Proc Natl Acad Sci U S A. 2012 Aug 14;109(33):13278-83. doi: 10.1073/pnas.1203536109. Epub 2012 Jul 30. Proc Natl Acad Sci U S A. 2012. PMID: 22847415 Free PMC article. - Functional replacement of the RING, B-box 2, and coiled-coil domains of tripartite motif 5alpha (TRIM5alpha) by heterologous TRIM domains.
Li X, Li Y, Stremlau M, Yuan W, Song B, Perron M, Sodroski J. Li X, et al. J Virol. 2006 Jul;80(13):6198-206. doi: 10.1128/JVI.00283-06. J Virol. 2006. PMID: 16775307 Free PMC article. - Impact of TRIM5α in vivo.
Nakayama EE, Shioda T. Nakayama EE, et al. AIDS. 2015 Sep 10;29(14):1733-43. doi: 10.1097/QAD.0000000000000812. AIDS. 2015. PMID: 26372380 Free PMC article. Review. - TRIM5 structure, HIV-1 capsid recognition, and innate immune signaling.
Grütter MG, Luban J. Grütter MG, et al. Curr Opin Virol. 2012 Apr;2(2):142-50. doi: 10.1016/j.coviro.2012.02.003. Epub 2012 Mar 5. Curr Opin Virol. 2012. PMID: 22482711 Free PMC article. Review.
Cited by
- Regulation of Epstein-Barr Virus Minor Capsid Protein BORF1 by TRIM5α.
Lin LT, Lu YS, Huang HH, Chen H, Hsu SW, Chang LK. Lin LT, et al. Int J Mol Sci. 2022 Dec 5;23(23):15340. doi: 10.3390/ijms232315340. Int J Mol Sci. 2022. PMID: 36499678 Free PMC article. - HIV Infection: Shaping the Complex, Dynamic, and Interconnected Network of the Cytoskeleton.
Cabrera-Rodríguez R, Pérez-Yanes S, Lorenzo-Sánchez I, Trujillo-González R, Estévez-Herrera J, García-Luis J, Valenzuela-Fernández A. Cabrera-Rodríguez R, et al. Int J Mol Sci. 2023 Aug 23;24(17):13104. doi: 10.3390/ijms241713104. Int J Mol Sci. 2023. PMID: 37685911 Free PMC article. Review. - TRIM34 restricts HIV-1 and SIV capsids in a TRIM5α-dependent manner.
Ohainle M, Kim K, Komurlu Keceli S, Felton A, Campbell E, Luban J, Emerman M. Ohainle M, et al. PLoS Pathog. 2020 Apr 13;16(4):e1008507. doi: 10.1371/journal.ppat.1008507. eCollection 2020 Apr. PLoS Pathog. 2020. PMID: 32282853 Free PMC article. - Rhesus monkey TRIM5α protein SPRY domain contributes to AP-1 activation.
Na L, Tang YD, Wang C, Liu C, Wang X. Na L, et al. J Biol Chem. 2018 Feb 23;293(8):2661-2674. doi: 10.1074/jbc.RA117.000127. Epub 2017 Dec 1. J Biol Chem. 2018. PMID: 29196608 Free PMC article. - K63-Linked Ubiquitin Is Required for Restriction of HIV-1 Reverse Transcription and Capsid Destabilization by Rhesus TRIM5α.
Imam S, Kömürlü S, Mattick J, Selyutina A, Talley S, Eddins A, Diaz-Griffero F, Campbell EM. Imam S, et al. J Virol. 2019 Jun 28;93(14):e00558-19. doi: 10.1128/JVI.00558-19. Print 2019 Jul 15. J Virol. 2019. PMID: 31068426 Free PMC article.
References
- Best S, Le Tissier P, Towers G, Stoye JP. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382:826–829. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical