Ribosome profiling reveals post-transcriptional buffering of divergent gene expression in yeast - PubMed (original) (raw)
Ribosome profiling reveals post-transcriptional buffering of divergent gene expression in yeast
C Joel McManus et al. Genome Res. 2014 Mar.
Abstract
Understanding the patterns and causes of phenotypic divergence is a central goal in evolutionary biology. Much work has shown that mRNA abundance is highly variable between closely related species. However, the extent and mechanisms of post-transcriptional gene regulatory evolution are largely unknown. Here we used ribosome profiling to compare transcript abundance and translation efficiency in two closely related yeast species (S. cerevisiae and S. paradoxus). By comparing translation regulatory divergence to interspecies differences in mRNA sequence features, we show that differences in transcript leaders and codon bias substantially contribute to divergent translation. Globally, we find that translation regulatory divergence often buffers species differences in mRNA abundance, such that ribosome occupancy is more conserved than transcript abundance. We used allele-specific ribosome profiling in interspecies hybrids to compare the relative contributions of cis- and trans-regulatory divergence to species differences in mRNA abundance and translation efficiency. The mode of gene regulatory divergence differs for these processes, as trans-regulatory changes play a greater role in divergent mRNA abundance than in divergent translation efficiency. Strikingly, most genes with aberrant transcript abundance in F1 hybrids (either over- or underexpressed compared to both parent species) did not exhibit aberrant ribosome occupancy. Our results show that interspecies differences in translation contribute substantially to the evolution of gene expression. Compensatory differences in transcript abundance and translation efficiency may increase the robustness of gene regulation.
Figures
Figure 1.
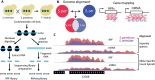
Overview of allele-specific ribosome profiling to measure divergence in ribosome occupancy, mRNA abundance, and translational efficiency. (A) Ribosome profiling was performed on log-phase cultures of S. cerevisiae, S. paradoxus, and their F1 hybrid. Ribosome protected fragments (RPFs) were purified and cloned into Illumina high-throughput sequencing libraries (left). Poly-adenylated mRNA sequencing libraries were prepared in parallel (right). (B) Sequence reads from each sample were aligned to both species genomes. Allele-specific reads were identified by comparing genomic alignments and mapped to corresponding regions of orthologous ORFs. (C) Comparison of separate and allele-specific coverage in ribosome profiling experiments. IGV browser tracks showing normalized coverage of RPF and mRNA sequence reads from S. cerevisiae (blue) and S. paradoxus (magenta) over the COX6 gene (YHR051W). Measurements of interspecies differences in read coverage are equivalent for separate species (upper) and allele-specific alignments (“mock hybrid,” lower) (see also Supplemental Fig. S1).
Figure 2.
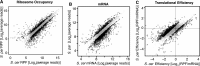
Comparing regulatory divergence of ribosome occupancy, mRNA abundance, and translation efficiency. (A–C) Scatter plots compare the normalized average number of sequence reads for S. cerevisiae (_x_-axis) and S. paradoxus (_y_-axis). Genes with statistically significant differences in read counts (FDR < 5%, minimum 1.5-fold difference) are plotted as open circles with black edges. Translation efficiency is defined here as the number of ribosome protected fragment reads (RPF) divided by the number of mRNA-seq reads covering an ORF.
Figure 3.
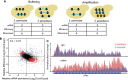
Translation regulatory divergence buffers interspecies differences in mRNA abundance. (A) Cartoon depicting buffering (left) and amplification (right). mRNA are shown as blue lines, and ribosomes are shown as black circles. Buffered genes have divergent mRNA abundance, with less divergent ribosome occupancy such that protein production is more conserved. In contrast, amplified genes have divergent mRNA abundance and even more divergent ribosome occupancy. (B) Scatterplot comparing divergent translation efficiency (_y_-axis, RPF/mRNA) with divergent mRNA abundance (_x_-axis). Buffered and amplified genes are plotted in red and blue, respectively. The negative correlation between mRNA abundance and translation efficiency suggests a genome-wide trend toward buffering. (C) Example of RPF and RNA-seq coverage over FPK1, a buffered gene. IGV browser tracks showing normalized coverage for S. cerevisiae (blue) and S. paradoxus (red).
Figure 4.
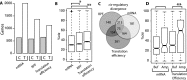
Contributions of _cis_- and _trans_-regulatory divergence in mRNA abundance, ribosome occupancy, and translation efficiency. (A) Bar plot shows the number of genes affected by significant regulatory divergence in _cis-_acting sequences (C) and _trans_-acting factors (T). _Trans_-acting factors contribute most to mRNA abundance. (B) Box plot showing the fraction of regulatory divergence attributable to differences in _cis_-regulatory elements (%cis). Ribosome occupancy and translation efficiency both have higher %cis than mRNA abundance. (C) Venn diagram showing the overlap of genes with _cis_-regulatory divergence in mRNA abundance, ribosome occupancy, and translation efficiency. (D) Box plot depicting %cis for genes with buffering (Buf) and amplifying (Amp) regulatory divergence of mRNA abundance (left) and translation efficiency (right). Asterisks indicate results of Wilcoxon rank-sum test comparisons: (*) P < 0.001; (**) P < 0.0005.
Figure 5.
Inheritance of gene expression in F1 hybrids of S. cerevisiae and S. paradoxus. (A) Hypothetical patterns of gene expression in S. paradoxus (red), S. cerevisiae (blue), and F1 hybrid yeast (purple), depicting six modes of gene expression inheritance. (B) Scatterplots showing the difference between mRNA expression levels (left) in the F1 hybrid and S. cerevisiae (_x_-axis) and S. paradoxus (_y_-axis). The difference between ribosome occupancy levels (RPF) in the F1 hybrid and parental species is shown on the right. Bar plots show the number of genes in each inheritance category from ribosome occupancy data. Genes with overdominant (overexpressed) and underdominant (underexpressed) inheritance occur much less frequently when considering ribosome occupancy, as compared to misexpression in mRNA abundance.
Similar articles
- Evolution at two levels of gene expression in yeast.
Artieri CG, Fraser HB. Artieri CG, et al. Genome Res. 2014 Mar;24(3):411-21. doi: 10.1101/gr.165522.113. Epub 2013 Dec 6. Genome Res. 2014. PMID: 24318729 Free PMC article. - Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling.
Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Ingolia NT, et al. Science. 2009 Apr 10;324(5924):218-23. doi: 10.1126/science.1168978. Epub 2009 Feb 12. Science. 2009. PMID: 19213877 Free PMC article. - Gene- and Species-Specific Hox mRNA Translation by Ribosome Expansion Segments.
Leppek K, Fujii K, Quade N, Susanto TT, Boehringer D, Lenarčič T, Xue S, Genuth NR, Ban N, Barna M. Leppek K, et al. Mol Cell. 2020 Dec 17;80(6):980-995.e13. doi: 10.1016/j.molcel.2020.10.023. Epub 2020 Nov 16. Mol Cell. 2020. PMID: 33202249 Free PMC article. - Recent advances in ribosome profiling for deciphering translational regulation.
Wang Y, Zhang H, Lu J. Wang Y, et al. Methods. 2020 Apr 1;176:46-54. doi: 10.1016/j.ymeth.2019.05.011. Epub 2019 May 17. Methods. 2020. PMID: 31103613 Review. - Regulation of fungal gene expression via short open reading frames in the mRNA 5'untranslated region.
Vilela C, McCarthy JE. Vilela C, et al. Mol Microbiol. 2003 Aug;49(4):859-67. doi: 10.1046/j.1365-2958.2003.03622.x. Mol Microbiol. 2003. PMID: 12890013 Review.
Cited by
- Negative feedback buffers effects of regulatory variants.
Bader DM, Wilkening S, Lin G, Tekkedil MM, Dietrich K, Steinmetz LM, Gagneur J. Bader DM, et al. Mol Syst Biol. 2015 Jan 29;11(1):785. doi: 10.15252/msb.20145844. Mol Syst Biol. 2015. PMID: 25634765 Free PMC article. - RiboGraph: An interactive visualization system for ribosome profiling data at read length resolution.
Chacko J, Ozadam H, Cenik C. Chacko J, et al. bioRxiv [Preprint]. 2024 Jan 12:2024.01.11.575228. doi: 10.1101/2024.01.11.575228. bioRxiv. 2024. PMID: 38260303 Free PMC article. Updated. Preprint. - Genomic variation. Impact of regulatory variation from RNA to protein.
Battle A, Khan Z, Wang SH, Mitrano A, Ford MJ, Pritchard JK, Gilad Y. Battle A, et al. Science. 2015 Feb 6;347(6222):664-7. doi: 10.1126/science.1260793. Epub 2014 Dec 18. Science. 2015. PMID: 25657249 Free PMC article. - Pichia pastoris regulates its gene-specific response to different carbon sources at the transcriptional, rather than the translational, level.
Prielhofer R, Cartwright SP, Graf AB, Valli M, Bill RM, Mattanovich D, Gasser B. Prielhofer R, et al. BMC Genomics. 2015 Mar 11;16(1):167. doi: 10.1186/s12864-015-1393-8. BMC Genomics. 2015. PMID: 25887254 Free PMC article.
References
- Benjamini Y, Hochberg Y 1995. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57: 289–300
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases