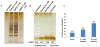Resin-assisted enrichment of thiols as a general strategy for proteomic profiling of cysteine-based reversible modifications - PubMed (original) (raw)
Resin-assisted enrichment of thiols as a general strategy for proteomic profiling of cysteine-based reversible modifications
Jia Guo et al. Nat Protoc. 2014 Jan.
Abstract
Reversible modifications of cysteine thiols have a key role in redox signaling and regulation. A number of reversible redox modifications, including disulfide formation, S-nitrosylation (SNO) and S-glutathionylation (SSG), have been recognized for their significance in various physiological and pathological processes. Here we describe a procedure for the enrichment of peptides containing reversible cysteine modifications. Starting with tissue or cell lysate samples, all of the unmodified free thiols are blocked using N-ethylmaleimide (NEM). This is followed by the selective reduction of those cysteines bearing the reversible modification(s) of interest. The reduction is achieved by using different reducing reagents that react specifically with each type of cysteine modification (e.g., ascorbate for SNO). This protocol serves as a general approach for enrichment of thiol-containing proteins or peptides derived from reversibly modified proteins. The approach uses a commercially available thiol-affinity resin (thiopropyl Sepharose 6B) to directly capture free thiol-containing proteins through a disulfide exchange reaction, followed by on-resin protein digestion and multiplexed isobaric labeling to facilitate liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based quantitative site-specific analysis of cysteine-based reversible modifications. The overall approach requires a simpler workflow with increased specificity compared with the commonly used biotinylation-based assays. The procedure for selective enrichment and analyses of SNO and the level of total reversible cysteine modifications (or total oxidation) is presented to demonstrate the utility of this general strategy. The entire protocol requires ∼3 d for sample processing with an additional day for LC-MS/MS and data analysis.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Figures
Figure 1
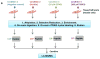
Schematic of the pre-processing and enrichment strategy for different reversible cysteine modifications. (a) Sample pre-processing strategies for different types of reversible cysteine modifications. The free thiols are initially blocked by alkylation and the different types of modified cysteines are selectively reduced to free thiols by using individual sets of reagents. Note that the term “total oxidation” is not exact as S-acylation is not a form of oxidation; however, the level of S-acylation is negligible compared to other forms of redox modifications. Also note that the breaking of S-acylation thioester bonds by DTT may not be effective since the reaction between DTT and thioesters is a much slower reaction compared to hydroxylamine. (b) Enrichment method for quantitative analysis of reversible cysteine modifications. After preprocessing, the free thiols derived from modified cysteines are captured by Thiopropyl Sepharose resin. Enriched proteins can be directly eluted for gel electrophoresis. On-resin digestion and on-resin isobaric labeling are performed for quantitative LC MS/MS analysis.
Figure 1
Schematic of the pre-processing and enrichment strategy for different reversible cysteine modifications. (a) Sample pre-processing strategies for different types of reversible cysteine modifications. The free thiols are initially blocked by alkylation and the different types of modified cysteines are selectively reduced to free thiols by using individual sets of reagents. Note that the term “total oxidation” is not exact as S-acylation is not a form of oxidation; however, the level of S-acylation is negligible compared to other forms of redox modifications. Also note that the breaking of S-acylation thioester bonds by DTT may not be effective since the reaction between DTT and thioesters is a much slower reaction compared to hydroxylamine. (b) Enrichment method for quantitative analysis of reversible cysteine modifications. After preprocessing, the free thiols derived from modified cysteines are captured by Thiopropyl Sepharose resin. Enriched proteins can be directly eluted for gel electrophoresis. On-resin digestion and on-resin isobaric labeling are performed for quantitative LC MS/MS analysis.
Figure 2
Enrichment of SNO-modified peptides from mouse muscle cells. (a) Experimental design and workflow with 4-plex iTRAQ labeling (114–117 m/z) for LC MS/MS analysis. (b) An MS/MS spectrum of the Cys-peptide #STEECLSYFGVSETTGLTPDQVK# from SERCA1 (UniProt:AT2A1_Mouse). The symbol # stands for the iTRAQ label on N-terminus and lysine. (c) The zoom-in spectrum of the reporter-ion region showing the increases of SNO levels on the identified peptide in response to GSNO treatment.
Figure 2
Enrichment of SNO-modified peptides from mouse muscle cells. (a) Experimental design and workflow with 4-plex iTRAQ labeling (114–117 m/z) for LC MS/MS analysis. (b) An MS/MS spectrum of the Cys-peptide #STEECLSYFGVSETTGLTPDQVK# from SERCA1 (UniProt:AT2A1_Mouse). The symbol # stands for the iTRAQ label on N-terminus and lysine. (c) The zoom-in spectrum of the reporter-ion region showing the increases of SNO levels on the identified peptide in response to GSNO treatment.
Figure 3
(a) SDS-gel image of enriched SNO-modified proteins from RAW 264.7 cells treated with CysNO for 15 min. UV exposure was performed in a 60 mm petri dish, 6 cm from a 15-watt UV lamp (λmax 365 nm) for 15 min. (b) SDS-gel image of enriched oxidized cysteine-containing peptides (the first 3 lanes) treated with diamide (0 mM, 0.1 mM and 0.5 mM) for 30 min and the total cysteine-containing peptides (lane 4) from RAW cells. (c) Stoichiometry of oxidation as measured by LC-MS/MS of the iTRAQ-labeled samples. Plotted here is the average stoichiometry across all identified peptides under different conditions. The stoichiometry of oxidation of each peptide was calculated as a percentage by dividing the reporter ion intensity for each channel (oxidized thiol) against the intensity of total thiol channel (enriched without NEM blocking).
Similar articles
- Proteomic identification and quantification of S-glutathionylation in mouse macrophages using resin-assisted enrichment and isobaric labeling.
Su D, Gaffrey MJ, Guo J, Hatchell KE, Chu RK, Clauss TR, Aldrich JT, Wu S, Purvine S, Camp DG, Smith RD, Thrall BD, Qian WJ. Su D, et al. Free Radic Biol Med. 2014 Feb;67:460-70. doi: 10.1016/j.freeradbiomed.2013.12.004. Epub 2013 Dec 11. Free Radic Biol Med. 2014. PMID: 24333276 Free PMC article. - Site-Specific Proteomic Mapping of Modified Cysteine Residues.
Gould NS. Gould NS. Methods Mol Biol. 2019;1967:183-195. doi: 10.1007/978-1-4939-9187-7_11. Methods Mol Biol. 2019. PMID: 31069771 - Solid-phase capture for the detection and relative quantification of S-nitrosoproteins by mass spectrometry.
Thompson JW, Forrester MT, Moseley MA, Foster MW. Thompson JW, et al. Methods. 2013 Aug 1;62(2):130-7. doi: 10.1016/j.ymeth.2012.10.001. Epub 2012 Oct 11. Methods. 2013. PMID: 23064468 Free PMC article. Review. - Large-scale capture of peptides containing reversibly oxidized cysteines by thiol-disulfide exchange applied to the myocardial redox proteome.
Paulech J, Solis N, Edwards AV, Puckeridge M, White MY, Cordwell SJ. Paulech J, et al. Anal Chem. 2013 Apr 2;85(7):3774-80. doi: 10.1021/ac400166e. Epub 2013 Mar 12. Anal Chem. 2013. PMID: 23438843 - Activity-Based Sensing for Site-Specific Proteomic Analysis of Cysteine Oxidation.
Shi Y, Carroll KS. Shi Y, et al. Acc Chem Res. 2020 Jan 21;53(1):20-31. doi: 10.1021/acs.accounts.9b00562. Epub 2019 Dec 23. Acc Chem Res. 2020. PMID: 31869209 Free PMC article. Review.
Cited by
- Sample multiplexing with cysteine-selective approaches: cysDML and cPILOT.
Gu L, Evans AR, Robinson RA. Gu L, et al. J Am Soc Mass Spectrom. 2015 Apr;26(4):615-30. doi: 10.1007/s13361-014-1059-9. Epub 2015 Jan 15. J Am Soc Mass Spectrom. 2015. PMID: 25588721 - Mass spectrometry-based proteomics for system-level characterization of biological responses to engineered nanomaterials.
Zhang T, Gaffrey MJ, Thrall BD, Qian WJ. Zhang T, et al. Anal Bioanal Chem. 2018 Sep;410(24):6067-6077. doi: 10.1007/s00216-018-1168-6. Epub 2018 Jun 8. Anal Bioanal Chem. 2018. PMID: 29947897 Free PMC article. Review. - Insights Into Protein _S_-Palmitoylation in Synaptic Plasticity and Neurological Disorders: Potential and Limitations of Methods for Detection and Analysis.
Zaręba-Kozioł M, Figiel I, Bartkowiak-Kaczmarek A, Włodarczyk J. Zaręba-Kozioł M, et al. Front Mol Neurosci. 2018 May 29;11:175. doi: 10.3389/fnmol.2018.00175. eCollection 2018. Front Mol Neurosci. 2018. PMID: 29910712 Free PMC article. Review. - Characterization of cellular oxidative stress response by stoichiometric redox proteomics.
Zhang T, Gaffrey MJ, Li X, Qian WJ. Zhang T, et al. Am J Physiol Cell Physiol. 2021 Feb 1;320(2):C182-C194. doi: 10.1152/ajpcell.00040.2020. Epub 2020 Dec 2. Am J Physiol Cell Physiol. 2021. PMID: 33264075 Free PMC article. Review. - Differential alkylation-based redox proteomics--Lessons learnt.
Wojdyla K, Rogowska-Wrzesinska A. Wojdyla K, et al. Redox Biol. 2015 Dec;6:240-252. doi: 10.1016/j.redox.2015.08.005. Epub 2015 Aug 5. Redox Biol. 2015. PMID: 26282677 Free PMC article. Review.
References
- Giron P, Dayon L, Sanchez JC. Cysteine tagging for MS-based proteomics. Mass Spectrom Rev. 2011;30:366–395. - PubMed
- Bachi A, Dalle-Donne I, Scaloni A. Redox proteomics: chemical principles, methodological approaches and biological/biomedical promises. Chem Rev. 2013;113:596–698. - PubMed
- Sato Y, Inaba K. Disulfide bond formation network in the three biological kingdoms, bacteria, fungi and mammals. FEBS J. 2012;279:2262–2271. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources