Cell cycle-dependent turnover of 5-hydroxymethyl cytosine in mouse embryonic stem cells - PubMed (original) (raw)
Cell cycle-dependent turnover of 5-hydroxymethyl cytosine in mouse embryonic stem cells
Junji Otani et al. PLoS One. 2013.
Abstract
Hydroxymethylcytosine in the genome is reported to be an intermediate of demethylation. In the present study, we demonstrated that maintenance methyltransferase Dnmt1 scarcely catalyzed hemi-hydroxymethylated DNA and that the hemi-hydroxymethylated DNA was not selectively recognized by the SRA domain of Uhrf1, indicating that hydroxymethylcytosine is diluted in a replication-dependent manner. A high level of 5-hydroxymethylcytosine in mouse embryonic stem cells was produced from the methylcytosine supplied mainly by de novo-type DNA methyltransferases Dnmt3a and Dnmt3b. The promoter regions of the HoxA gene cluster showed a high hydroxymethylation level whilst the methylcytosine level was quite low, suggesting that methylated CpG is actively hydroxylated during proliferation. All the results indicate that removal and production of hydroxymethylcytosine are regulated in replication-dependent manners in mouse embryonic stem cells.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
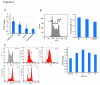
Figure 1. 5hmC content is diluted during replication.
A. Hemi-hydroxymethylated DNA (CG/5hmCG) is not a good substrate for Dnmt1. The DNA methylation activity of mouse Dnmt1, Dnmt3a, and Dnmt3b towards 35-bp unmethylated (CG/CG), hemi-methylated (CG/5mCG), or hemi-hydroxymethylated (CG/5hmCG) DNA was determined. B. Gel mobility shift assaying of the SRA domain of mouse Uhrf1. The indicated concentrations of SRA were incubated with either 12-bp CG/5mC, CG/5hmCG, or CG/CG, followed by electrophoresis (left panel). The complex of the SRA and 32P-labeled CG/5mCG was competed with the indicated amounts of non-labeled CG/5mCG, CG/5hmCG, or CG/CG DNA (right panel). DNA bound to SRA (B) and free DNA (F) are indicated.
Figure 2. Cell cycle-dependent change in the 5hmC content.
A. The 5hmC content in mESCs treated with aphidicolin, hydroxyurea, serum depletion, or nocodazole was determined by β-GT assaying. The values represent the fold change normalized as to that without treatment. The values for each treatment are averages ± SD (n=3). B. The 5hmC content in mESCs sorted by FACS (left panel) was determined (right panel). The values are averages ± SE (n=3). C. The non-synchronized (w/o S) and synchronized mESCs were collected after the indicated times and the 5hmC contents were determined. The left panels show the results of FACS analyses and the right panel the 5hmC content.
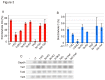
Figure 3. Dnmt3a and Dnmt3b mainly provide 5mC for the hydroxymethylation in mESCs.
The 5mC, 5hmC, and Tet mRNA contents of J1 parent, Dnmt1 (1-KO), Dnmt3a and Dnmt3b (3-DKO), Dnmt3a (3a-KO), Dnmt3b (3b-KO), and Dnmt1, Dnmt3a and Dnmt3b (TKO) knockout mESCs, and ectopically expressed TAP-tagged Dnmt3a (3a-TAP) or Dnmt3a2 (3a2-TAP) in 3-DKO mESCs were determined. A. The 5mC contents (%) were determined as M.SssI methylation ability from a standard curve (Figure S1A). B. The 5hmC contents were determined from the standard curve obtained on β-GT assaying (Figure S1B). The values are the averages ± SD determined for three independent genomic DNA samples. C. Relative mRNA expression of Tet1, Tet2, and Tet3 was evaluated by semi-quantitative RT-PCR. (-) indicates the product of PCR without the template.
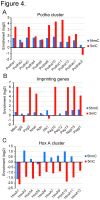
Figure 4. Enrichment of 5mC and 5hmC in specific promoters.
5hmC (blue bars) and 5mC (red bars) were determined by DNA microarray analysis in the promoters of the Pcdha gene cluster (A), maternal imprinting genes (B), and HoxA gene cluster (C). The abscissas indicate enrichment of 5hmC or 5mC on a log2 scale.
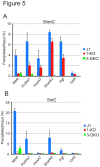
Figure 5. Dnmt3a and Dnmt3b-dependent 5mC are responsible for the production of 5hmC.
The 5hmC (A) and 5mC (B) contents of J1 (blue bars), Dnmt1 (1-KO, red bars), and Dnmt3a and Dnmt3b (3-DKO, light green bars) knockout mESCs were determined by q-PCR in the promoters of five representative 5hmC-enriched genes. The values are the averages + SD determined for three independent genomic DNA samples.
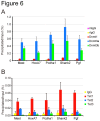
Figure 6. Dnmt1, Dnmt3a, Dnmt3b, and Tet1 are recruited to 5hmC-enriched promoters.
The occupancy of Dnmt1, Dnmt3a, and Dnmt3b (A), and Tet1, Tet2, and Tet3 (B) was determined by ChIP-qPCR in the promoters of the 5hmC-enriched genes shown in Figure 4. The values are the averages + SD determined for three independent DNA samples.
Similar articles
- UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9.
Liu X, Gao Q, Li P, Zhao Q, Zhang J, Li J, Koseki H, Wong J. Liu X, et al. Nat Commun. 2013;4:1563. doi: 10.1038/ncomms2562. Nat Commun. 2013. PMID: 23463006 - S phase-dependent interaction with DNMT1 dictates the role of UHRF1 but not UHRF2 in DNA methylation maintenance.
Zhang J, Gao Q, Li P, Liu X, Jia Y, Wu W, Li J, Dong S, Koseki H, Wong J. Zhang J, et al. Cell Res. 2011 Dec;21(12):1723-39. doi: 10.1038/cr.2011.176. Epub 2011 Nov 8. Cell Res. 2011. PMID: 22064703 Free PMC article. - The DNA methyltransferase Dnmt1 directly interacts with the SET and RING finger-associated (SRA) domain of the multifunctional protein Uhrf1 to facilitate accession of the catalytic center to hemi-methylated DNA.
Berkyurek AC, Suetake I, Arita K, Takeshita K, Nakagawa A, Shirakawa M, Tajima S. Berkyurek AC, et al. J Biol Chem. 2014 Jan 3;289(1):379-86. doi: 10.1074/jbc.M113.523209. Epub 2013 Nov 19. J Biol Chem. 2014. PMID: 24253042 Free PMC article. - Regulation of maintenance DNA methylation via histone ubiquitylation.
Nishiyama A, Yamaguchi L, Nakanishi M. Nishiyama A, et al. J Biochem. 2016 Jan;159(1):9-15. doi: 10.1093/jb/mvv113. Epub 2015 Nov 20. J Biochem. 2016. PMID: 26590302 Free PMC article. Review. - Coordinated Dialogue between UHRF1 and DNMT1 to Ensure Faithful Inheritance of Methylated DNA Patterns.
Bronner C, Alhosin M, Hamiche A, Mousli M. Bronner C, et al. Genes (Basel). 2019 Jan 18;10(1):65. doi: 10.3390/genes10010065. Genes (Basel). 2019. PMID: 30669400 Free PMC article. Review.
Cited by
- A novel pax5-binding regulatory element in the igκ locus.
Levin-Klein R, Kirillov A, Rosenbluh C, Cedar H, Bergman Y. Levin-Klein R, et al. Front Immunol. 2014 May 23;5:240. doi: 10.3389/fimmu.2014.00240. eCollection 2014. Front Immunol. 2014. PMID: 24904588 Free PMC article. - Dysregulation of the TET family of epigenetic regulators in lymphoid and myeloid malignancies.
Lio CJ, Yuita H, Rao A. Lio CJ, et al. Blood. 2019 Oct 31;134(18):1487-1497. doi: 10.1182/blood.2019791475. Blood. 2019. PMID: 31467060 Free PMC article. Review. - Reversing DNA methylation: mechanisms, genomics, and biological functions.
Wu H, Zhang Y. Wu H, et al. Cell. 2014 Jan 16;156(1-2):45-68. doi: 10.1016/j.cell.2013.12.019. Cell. 2014. PMID: 24439369 Free PMC article. - Generation of new hair cells by DNA methyltransferase (Dnmt) inhibitor 5-azacytidine in a chemically-deafened mouse model.
Deng X, Liu Z, Li X, Zhou Y, Hu Z. Deng X, et al. Sci Rep. 2019 May 29;9(1):7997. doi: 10.1038/s41598-019-44313-0. Sci Rep. 2019. PMID: 31142766 Free PMC article. - TET Methylcytosine Oxidases in T Cell and B Cell Development and Function.
Tsagaratou A, Lio CJ, Yue X, Rao A. Tsagaratou A, et al. Front Immunol. 2017 Mar 31;8:220. doi: 10.3389/fimmu.2017.00220. eCollection 2017. Front Immunol. 2017. PMID: 28408905 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources