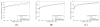Fast and accurate database searches with MS-GF+Percolator - PubMed (original) (raw)
. 2014 Feb 7;13(2):890-7.
doi: 10.1021/pr400937n. Epub 2013 Dec 23.
Affiliations
- PMID: 24344789
- PMCID: PMC3975676
- DOI: 10.1021/pr400937n
Fast and accurate database searches with MS-GF+Percolator
Viktor Granholm et al. J Proteome Res. 2014.
Abstract
One can interpret fragmentation spectra stemming from peptides in mass-spectrometry-based proteomics experiments using so-called database search engines. Frequently, one also runs post-processors such as Percolator to assess the confidence, infer unique peptides, and increase the number of identifications. A recent search engine, MS-GF+, has shown promising results, due to a new and efficient scoring algorithm. However, MS-GF+ provides few statistical estimates about the peptide-spectrum matches, hence limiting the biological interpretation. Here, we enabled Percolator processing for MS-GF+ output and observed an increased number of identified peptides for a wide variety of data sets. In addition, Percolator directly reports p values and false discovery rate estimates, such as q values and posterior error probabilities, for peptide-spectrum matches, peptides, and proteins, functions that are useful for the whole proteomics community.
Conflict of interest statement
Notes
The authors declare no conflicting interests.
Figures
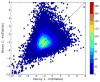
Figure 1. The covariation of two MS-GF+ decoy searches
MS-GF+ was run against a reversed decoy database (Decoy 1) and a shuffled decoy database (Decoy 2) generated with Mimic. For each PSM, the negative logarithm of the E value reported by MS-GF+ was plotted.
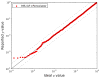
Figure 2. Statistical calibration of null p values from MS-GF+Percolator
Using only incorrect matches from the Orbitrap spectra of ISB18 mix 7, p values were estimated from the Percolator score, after post-processing the MS-GF+ results. The y axis represents the p values reported by Percolator, and the x axis a uniform rank from 0 to 1. The black line marks the x = y diagonal, and the gray dashed lines x = 2_y_ and x = y/2. A calculated Kolomogorov-Smirnov test D value of 0.014 between the reported and ideal p values indicates a small difference between the two distributions.
Figure 3. MS-GF+Percolator performance on tryptic datasets
The numbers of accepted unique peptides are plotted as a function of the peptide level q value threshold. Panel (A) shows the results for the human prostate cancer data, (B) the phosphorylated mouse data and (C) the TMT labeled iPRG data. Peptide level confidence estimates from MS-GF+ were obtained by using qvality as described in Experimental procedures.
Figure 4. The performance of MS-GF+Percolator on the yeast data digested by multiple enzymes
The number of accepted unique peptides for different peptide level q values thresholds, for several different enzymes. The enzymes used for digestions are (A) Arg-C, (B) Asp-N, (C) Glu-C, (D) Lys-C and (E) trypsin.) Peptide level confidence estimates from MS-GF+ were obtained by using qvality as described in Experimental procedures.
Similar articles
- Fast and Accurate Protein False Discovery Rates on Large-Scale Proteomics Data Sets with Percolator 3.0.
The M, MacCoss MJ, Noble WS, Käll L. The M, et al. J Am Soc Mass Spectrom. 2016 Nov;27(11):1719-1727. doi: 10.1007/s13361-016-1460-7. Epub 2016 Aug 29. J Am Soc Mass Spectrom. 2016. PMID: 27572102 Free PMC article. - Optimization of Search Engines and Postprocessing Approaches to Maximize Peptide and Protein Identification for High-Resolution Mass Data.
Tu C, Sheng Q, Li J, Ma D, Shen X, Wang X, Shyr Y, Yi Z, Qu J. Tu C, et al. J Proteome Res. 2015 Nov 6;14(11):4662-73. doi: 10.1021/acs.jproteome.5b00536. Epub 2015 Sep 30. J Proteome Res. 2015. PMID: 26390080 Free PMC article. - Enhanced peptide identification by electron transfer dissociation using an improved Mascot Percolator.
Wright JC, Collins MO, Yu L, Käll L, Brosch M, Choudhary JS. Wright JC, et al. Mol Cell Proteomics. 2012 Aug;11(8):478-91. doi: 10.1074/mcp.O111.014522. Epub 2012 Apr 6. Mol Cell Proteomics. 2012. PMID: 22493177 Free PMC article. - Modification Site Localization in Peptides.
Chalkley RJ. Chalkley RJ. Adv Exp Med Biol. 2016;919:243-247. doi: 10.1007/978-3-319-41448-5_13. Adv Exp Med Biol. 2016. PMID: 27975222 Review. - A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics.
Nesvizhskii AI. Nesvizhskii AI. J Proteomics. 2010 Oct 10;73(11):2092-123. doi: 10.1016/j.jprot.2010.08.009. Epub 2010 Sep 8. J Proteomics. 2010. PMID: 20816881 Free PMC article. Review.
Cited by
- Proteome Analysis of Pancreatic Tumors Implicates Extracellular Matrix in Patient Outcome.
Silwal-Pandit L, Stålberg SM, Johansson HJ, Mermelekas G, Lothe IMB, Skrede ML, Dalsgaard AM, Nebdal DJH, Helland Å, Lingjærde OC, Labori KJ, Skålhegg BS, Lehtiö J, Kure EH. Silwal-Pandit L, et al. Cancer Res Commun. 2022 Jun 14;2(6):434-446. doi: 10.1158/2767-9764.CRC-21-0100. eCollection 2022 Jun. Cancer Res Commun. 2022. PMID: 36923555 Free PMC article. - TIDD: tool-independent and data-dependent machine learning for peptide identification.
Li H, Na S, Hwang KB, Paek E. Li H, et al. BMC Bioinformatics. 2022 Mar 30;23(1):109. doi: 10.1186/s12859-022-04640-y. BMC Bioinformatics. 2022. PMID: 35354356 Free PMC article. - Annotating N termini for the human proteome project: N termini and Nα-acetylation status differentiate stable cleaved protein species from degradation remnants in the human erythrocyte proteome.
Lange PF, Huesgen PF, Nguyen K, Overall CM. Lange PF, et al. J Proteome Res. 2014 Apr 4;13(4):2028-44. doi: 10.1021/pr401191w. Epub 2014 Mar 10. J Proteome Res. 2014. PMID: 24555563 Free PMC article. - Sipros Ensemble improves database searching and filtering for complex metaproteomics.
Guo X, Li Z, Yao Q, Mueller RS, Eng JK, Tabb DL, Hervey WJ 4th, Pan C. Guo X, et al. Bioinformatics. 2018 Mar 1;34(5):795-802. doi: 10.1093/bioinformatics/btx601. Bioinformatics. 2018. PMID: 29028897 Free PMC article. - Improving protein identification from tandem mass spectrometry data by one-step methods and integrating data from other platforms.
Sikdar S, Gill R, Datta S. Sikdar S, et al. Brief Bioinform. 2016 Mar;17(2):262-9. doi: 10.1093/bib/bbv043. Epub 2015 Jul 3. Brief Bioinform. 2016. PMID: 26141827 Free PMC article. Review.
References
- Eng J, McCormack A, Yates J, et al. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. Journal of the American Society for Mass Spectrometry. 1994;5:976–989. - PubMed
- Perkins D, Pappin D, Creasy D, Cottrell J. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. - PubMed
- Craig R, Beavis R. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004:921. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources