The complete genome sequence of a Neanderthal from the Altai Mountains - PubMed (original) (raw)
Comparative Study
. 2014 Jan 2;505(7481):43-9.
doi: 10.1038/nature12886. Epub 2013 Dec 18.
Fernando Racimo 2, Nick Patterson 3, Flora Jay 2, Sriram Sankararaman 4, Susanna Sawyer 1, Anja Heinze 1, Gabriel Renaud 1, Peter H Sudmant 5, Cesare de Filippo 1, Heng Li 3, Swapan Mallick 4, Michael Dannemann 1, Qiaomei Fu 6, Martin Kircher 7, Martin Kuhlwilm 1, Michael Lachmann 1, Matthias Meyer 1, Matthias Ongyerth 1, Michael Siebauer 1, Christoph Theunert 1, Arti Tandon 4, Priya Moorjani 8, Joseph Pickrell 8, James C Mullikin 9, Samuel H Vohr 10, Richard E Green 10, Ines Hellmann 11, Philip L F Johnson 12, Hélène Blanche 13, Howard Cann 13, Jacob O Kitzman 5, Jay Shendure 5, Evan E Eichler 14, Ed S Lein 15, Trygve E Bakken 15, Liubov V Golovanova 16, Vladimir B Doronichev 16, Michael V Shunkov 17, Anatoli P Derevianko 17, Bence Viola 18, Montgomery Slatkin 2, David Reich 19, Janet Kelso 1, Svante Pääbo 1
Affiliations
- PMID: 24352235
- PMCID: PMC4031459
- DOI: 10.1038/nature12886
Comparative Study
The complete genome sequence of a Neanderthal from the Altai Mountains
Kay Prüfer et al. Nature. 2014.
Abstract
We present a high-quality genome sequence of a Neanderthal woman from Siberia. We show that her parents were related at the level of half-siblings and that mating among close relatives was common among her recent ancestors. We also sequenced the genome of a Neanderthal from the Caucasus to low coverage. An analysis of the relationships and population history of available archaic genomes and 25 present-day human genomes shows that several gene flow events occurred among Neanderthals, Denisovans and early modern humans, possibly including gene flow into Denisovans from an unknown archaic group. Thus, interbreeding, albeit of low magnitude, occurred among many hominin groups in the Late Pleistocene. In addition, the high-quality Neanderthal genome allows us to establish a definitive list of substitutions that became fixed in modern humans after their separation from the ancestors of Neanderthals and Denisovans.
Conflict of interest statement
Competing financial interests
The authors declare no competing financial interests.
Figures
Figure 1. Toe phalanx and location of Neandertal samples for which genome-wide data are available
a, The toe phalanx found in the East Gallery of Denisova Cave in 2010. Left: dorsal view; Right: left view. b, Map of Eurasia showing the location of Vindija cave, Mezmaiskaya cave and Denisova cave.
Figure 2. Phylogenetic relationships of the Altai Neandertal
a, Bayesian tree of mitochondrial sequences of the toe phalanx, the Denisovan finger phalanx, six Neandertals, and five present-day humans. Posterior probabilities are given for branches whose support is less than one (SI 2b). b, Neighbor-joining tree based on autosomal transversion differences among the toe phalanx, four Neandertals, the Denisova genome, and seven present-day human individuals. Bootstrap values are shown for branches supported by less than 100% of 1,000 bootstrap replicates (SI 6).
Figure 3. Indications of inbreeding in the Altai Neandertal individual
a, Time since the most recent common ancestor in log-scale for the two alleles of a French, the Denisovan and the Altai Neandertal individual (SI 12) along 40 Mb of chromosome 21. b, Pedigrees showing four possible scenarios of parental relatedness for the Altai Neandertal (i.e. the child at the bottom of each pedigree). Two additional scenarios can be derived by switching the sex of the parents for the panels marked with an asterisk. c, Fraction of the genome in runs of homozygosity between 2.5 and 10cM in length for Altai Neandertal, Denisovan and the three present-day human individuals with the largest fractions (grey bars). The fractions for the Altai Neandertal (bottom four bars) are reduced by the fraction expected from the four inbreeding scenarios in a.
Figure 4. Inference of population size change over time
The y-axis specifies a number proportional to the population size Ne. The x-axis specifies time in units of divergence per base pair (along the top in years for mutation rates of 0.5 × 10−9 to 1.0 × 10−9 per site per year). The analysis assumes that the Neandertal and Denisova remains are of the same age, whereas archaeological evidence and the branch shortening suggest that the Neandertal bone is older than the Denisovan bone. However, because the exact difference in ages is not known, it is not possible to determine whether the reduction in population size experienced by both archaic groups (but not by modern humans) coincided in time.
Figure 5. Relatedness of introgressing archaic and sequenced archaic samples
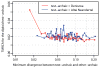
Divergence of phased present-day human genomes to archaic genomes in windows of size 0.01cM with a minimum of 25,000 analysed bases. Windows are sorted by sequence divergence measured on the archaic side of the tree (SI 13) and the y-axis reports the divergence relative to human-chimpanzee divergence for cumulative fractions of the sorted windows over the entire genomes. Regions of low divergence between non-Africans and Neandertals (a) and between Oceanians and Denisovans (b) indicate gene flow between these groups and the relative divergences between the introgressing archaic and sequenced archaic samples.
Figure 6. Neandertal gene flow into Siberian Denisovans
Divergence in 0.01cM sized windows with at least 50kb analyzed bases between a “test”-archaic genome and effectively haploid regions of the other archaic genome archaic plotted against the most recent-common ancestor of the two alleles of the “test”-archaic. The plot shows 50 equally sized bins of windows for the “test”-archaic Denisovan against the effectively haploid Neandertal (red) and for the “test”-archaic Altai Neandertal against the effectively haploid Denisovan (blue). Divergence is given as percentage of human-chimpanzee divergence. Windows that show a close relationship between the effective haploid Altai Neandertal and the closest inferred Denisovan haplotype show a deep divergence to the second Denisovan haplotype, indicating gene flow from Neandertal into Denisovan.
Figure 7. Altai and Denisovan allele sharing with Africans stratified by African allele frequency
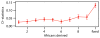
The plot shows the _D_-statistic of the form D(Neandertal, Denisova; Africa, Chimpanzee) binned by derived allele count in 10 deeply sequenced African genomes. Error-bars represent ± 1 standard error. High-frequency and fixed derived alleles in Africa are more often shared with the Neandertal than with Denisovan genome.
Figure 8. A possible model of gene flow events in the late Pleistocene
The direction and estimated magnitude of inferred gene flow events are shown. Branch lengths and ages gene flows are not drawn to scale. The dashed line indicates that it is uncertain if Denisovan gene flow into modern humans occurred once or more times. D.I. denotes the introgressing Denisovan, N.I. the introgressing Neandertal. Note that the age of the archaic genomes precludes detection of gene-flow from modern humans into the archaic hominins.
Extended Data Figure 1
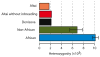
Heterozygosity estimates for the Altai Neandertal individual, the Denisovan individual, non-Africans and Africans. The bars for the latter two give the range of heterozygosity observed among 15 non-African and 10 African individuals, respectively (SI 9).
Extended Data Figure 2
Neandertal-introgressed loci in Denisova Divergence of the Altai Neandertal to the most closely related Denisovan haplotype in windows of at least 200kb on chromosome 6. Divergence is given as percentage of human-chimpanzee divergence and bars represent ± 1 standard error.
Comment in
- Archaic humans: Four makes a party.
Birney E, Pritchard JK. Birney E, et al. Nature. 2014 Jan 2;505(7481):32-4. doi: 10.1038/nature12847. Epub 2013 Dec 18. Nature. 2014. PMID: 24352230 No abstract available.
Similar articles
- Ancient gene flow from early modern humans into Eastern Neanderthals.
Kuhlwilm M, Gronau I, Hubisz MJ, de Filippo C, Prado-Martinez J, Kircher M, Fu Q, Burbano HA, Lalueza-Fox C, de la Rasilla M, Rosas A, Rudan P, Brajkovic D, Kucan Ž, Gušic I, Marques-Bonet T, Andrés AM, Viola B, Pääbo S, Meyer M, Siepel A, Castellano S. Kuhlwilm M, et al. Nature. 2016 Feb 25;530(7591):429-33. doi: 10.1038/nature16544. Epub 2016 Feb 17. Nature. 2016. PMID: 26886800 Free PMC article. - Reconstructing the genetic history of late Neanderthals.
Hajdinjak M, Fu Q, Hübner A, Petr M, Mafessoni F, Grote S, Skoglund P, Narasimham V, Rougier H, Crevecoeur I, Semal P, Soressi M, Talamo S, Hublin JJ, Gušić I, Kućan Ž, Rudan P, Golovanova LV, Doronichev VB, Posth C, Krause J, Korlević P, Nagel S, Nickel B, Slatkin M, Patterson N, Reich D, Prüfer K, Meyer M, Pääbo S, Kelso J. Hajdinjak M, et al. Nature. 2018 Mar 29;555(7698):652-656. doi: 10.1038/nature26151. Epub 2018 Mar 21. Nature. 2018. PMID: 29562232 Free PMC article. - Early history of Neanderthals and Denisovans.
Rogers AR, Bohlender RJ, Huff CD. Rogers AR, et al. Proc Natl Acad Sci U S A. 2017 Sep 12;114(37):9859-9863. doi: 10.1073/pnas.1706426114. Epub 2017 Aug 7. Proc Natl Acad Sci U S A. 2017. PMID: 28784789 Free PMC article. - Archaic human genomics.
Disotell TR. Disotell TR. Am J Phys Anthropol. 2012;149 Suppl 55:24-39. doi: 10.1002/ajpa.22159. Epub 2012 Nov 2. Am J Phys Anthropol. 2012. PMID: 23124308 Review. - Archaic hominin introgression into modern human genomes.
Gokcumen O. Gokcumen O. Am J Phys Anthropol. 2020 May;171 Suppl 70:60-73. doi: 10.1002/ajpa.23951. Epub 2019 Nov 8. Am J Phys Anthropol. 2020. PMID: 31702050 Review.
Cited by
- It's more than stamp collecting: how genome sequencing can unify biological research.
Richards S. Richards S. Trends Genet. 2015 Jul;31(7):411-21. doi: 10.1016/j.tig.2015.04.007. Epub 2015 May 20. Trends Genet. 2015. PMID: 26003218 Free PMC article. Review. - HuConTest: Testing Human Contamination in Great Ape Samples.
Kuhlwilm M, Fontsere C, Han S, Alvarez-Estape M, Marques-Bonet T. Kuhlwilm M, et al. Genome Biol Evol. 2021 Jun 8;13(6):evab117. doi: 10.1093/gbe/evab117. Genome Biol Evol. 2021. PMID: 34038549 Free PMC article. - Nuclear genetic diversity of head lice sheds light on human dispersal around the world.
Ascunce MS, Toloza AC, González-Oliver A, Reed DL. Ascunce MS, et al. PLoS One. 2023 Nov 8;18(11):e0293409. doi: 10.1371/journal.pone.0293409. eCollection 2023. PLoS One. 2023. PMID: 37939041 Free PMC article. - A genetic method for dating ancient genomes provides a direct estimate of human generation interval in the last 45,000 years.
Moorjani P, Sankararaman S, Fu Q, Przeworski M, Patterson N, Reich D. Moorjani P, et al. Proc Natl Acad Sci U S A. 2016 May 17;113(20):5652-7. doi: 10.1073/pnas.1514696113. Epub 2016 May 2. Proc Natl Acad Sci U S A. 2016. PMID: 27140627 Free PMC article. - Neanderthal introgression reintroduced functional ancestral alleles lost in Eurasian populations.
Rinker DC, Simonti CN, McArthur E, Shaw D, Hodges E, Capra JA. Rinker DC, et al. Nat Ecol Evol. 2020 Oct;4(10):1332-1341. doi: 10.1038/s41559-020-1261-z. Epub 2020 Jul 27. Nat Ecol Evol. 2020. PMID: 32719451 Free PMC article.
References
- Mednikova MB. A proximal pedal phalanx of a paleolithic hominin from Denisova cave, Altai. Archaeology Ethnology & Anthropology of Eurasia. 2011;39:129–138. http://dx.doi.org/10.1016/j.aeae.2011.06.017. - DOI
Publication types
MeSH terms
Grants and funding
- R01-GM40282/GM/NIGMS NIH HHS/United States
- R01 GM100233/GM/NIGMS NIH HHS/United States
- R01 GM040282/GM/NIGMS NIH HHS/United States
- HG002385/HG/NHGRI NIH HHS/United States
- HG006283/HG/NHGRI NIH HHS/United States
- GM100233/GM/NIGMS NIH HHS/United States
- R01 HG006283/HG/NHGRI NIH HHS/United States
- R01 HG002385/HG/NHGRI NIH HHS/United States