MIR376A is a regulator of starvation-induced autophagy - PubMed (original) (raw)
MIR376A is a regulator of starvation-induced autophagy
Gozde Korkmaz et al. PLoS One. 2013.
Erratum in
- PLoS One. 2014;9(7):e103385
Abstract
Background: Autophagy is a vesicular trafficking process responsible for the degradation of long-lived, misfolded or abnormal proteins, as well as damaged or surplus organelles. Abnormalities of the autophagic activity may result in the accumulation of protein aggregates, organelle dysfunction, and autophagy disorders were associated with various diseases. Hence, mechanisms of autophagy regulation are under exploration.
Methods: Over-expression of hsa-miR-376a1 (shortly MIR376A) was performed to evaluate its effects on autophagy. Autophagy-related targets of the miRNA were predicted using Microcosm Targets and MIRanda bioinformatics tools and experimentally validated. Endogenous miRNA was blocked using antagomirs and the effects on target expression and autophagy were analyzed. Luciferase tests were performed to confirm that 3' UTR sequences in target genes were functional. Differential expression of MIR376A and the related MIR376B was compared using TaqMan quantitative PCR.
Results: Here, we demonstrated that, a microRNA (miRNA) from the DLK1/GTL2 gene cluster, MIR376A, played an important role in autophagy regulation. We showed that, amino acid and serum starvation-induced autophagy was blocked by MIR376A overexpression in MCF-7 and Huh7 cells. MIR376A shared the same seed sequence and had overlapping targets with MIR376B, and similarly blocked the expression of key autophagy proteins ATG4C and BECN1 (Beclin 1). Indeed, 3' UTR sequences in the mRNA of these autophagy proteins were responsive to MIR376A in luciferase assays. Antagomir tests showed that, endogenous MIR376A was participating to the control of ATG4C and BECN1 transcript and protein levels. Moreover, blockage of endogenous MIR376A accelerated starvation-induced autophagic activity. Interestingly, MIR376A and MIR376B levels were increased with different kinetics in response to starvation stress and tissue-specific level differences were also observed, pointing out to an overlapping but miRNA-specific biological role.
Conclusions: Our findings underline the importance of miRNAs encoded by the DLK1/GTL2 gene cluster in stress-response control mechanisms, and introduce MIR376A as a new regulator of autophagy.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. MIR376 family miRNAs in the DLKI/GTL2 gene cluster.
(A) Individual miRNA genes were indicated. Sizes of spacer sequences between miRNA genes were marked as nucleotides (nt). (B) Comparison of mature miRNA sequences with identical “seed sequences” (bold, underlined). Identical nucleotides were indicated by asterix (*). ClustalW tool was used for the analysis. (C) ClustalW comparison of the stem-loop sequences of MIR376A and MIR376B. Seed sequences were marked as bold and underlined.
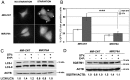
Figure 2. Effect of MIR376A overexpression on autophagy.
(A) MCF-7 cells were co-transfected with either MIR-CNT (control plasmid) or MIR376A and GFP-LC3 plasmid, and GFP-LC3 dot formation was analyzed. White arrows indicate clusters of the GFP-LC3 dots in cells. (B) Quantification of the experiments in A. MIR376A overexpression, but not MIR-CNT expression, blocked starvation (STV)-induced autophagy (mean ± S.D. of independent experiments, n = 4, ***p<0,01). NON STV, non-starved (C) Overexpression of MIR376A resulted in a decrease in the autophagic activity of MCF-7 cells. Starvation-induced conversion of LC3-I to LC3-II in MCF-7 cells was analyzed. Tests were performed in the presence or absence of E64d (10 µg/ml) and Pepstatin A (10 µg/ml) (E+P). LC3-II/LC3-I densitometric ratios were marked. ACTB was used as a loading control. (D) MIR376A blocked starvation induced SQSTM1 degradation in MCF-7 cells. ACTB was used as a loading control. SQSTM1/ACTIN densitometric ratios were marked.
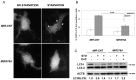
Figure 3. MIR376A overexpression blocked autophagy in Huh-7 cells.
(A) MIR376A blocked GFP-LC3 dot formation under starvation condition. (B) Quantitative analysis of experiments in A (mean ± S.D. of independent experiments, n = 3, ***p<0,01). (C) Overexpression of MIR376A resulted in decreased autophagic flux in Huh-7 cells. Starvation-induced conversion of LC3-I to LC3-II was analyzed. Tests were performed in the presence or absence of E64d (10 µg/ml) and Pepstatin A (10 µg/ml) (E+P). LC3-II/LC3-I densitometric ratios were marked. ACTB was used as a loading control.
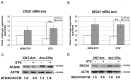
Figure 4. ATG4C and BECN1 were targets of MIR376A.
(A and B) MIR376A mature miRNA sequence and MIR376A binding target sequences in the 3' UTR of ATG4C (A) and BECN1(B) mRNAs were depicted. MIR376A seed sequence was shown in bold letters and and complemetary sequences were indicated. (C and D) qPCR analysis of ATG4C and BECN1 mRNA expression in control miRNA (MIR-CONT) or MIR376A overexpressing cells under non-starved (NON STV) or starved (STV) condition (mean ± S.D. of independent experiments, n = 3, *p<0,05). (E and F) Immunoblots showing ATG4C and BECN1 protein levels following control miRNA (MIR-CONT) or MIR376A overexpression. ACTB was used as a loading control. ATG4C/ACTB or BECN1/ACTB densitometric ratios were marked.
Figure 5. Effect of antagomirs on MIR376A target expression.
(A and B) qPCR analysis of ATG4C and BECN1 mRNA levels in MCF-7 cells transfected with control antagomirs (CNT-Ant) or antagomir-376a (Ant-376a) (mean ± S.D. of independent experiments, n = 3, **p<0,03 for ATG4C, and n = 3, **p<0,03 for BECN1). Results were expressed as fold changes against GAPDH mRNA levels. (C and D) ATG4C and BECN1 protein levels were increased following antagomir-376a (Ant-376a) transfection. ATG4C/ACTB or BECN1/ACTB densitometric ratios were marked.
Figure 6. MIR376A controlled miRNA-response elements (MREs) in ATG4C and BECN1 mRNA 3′ UTRs.
Schemes showing ATG4C (A) and (B) BECN1 MRE (top), or their mutated versions (bottom) placed in the 3' UTR of the luciferase gene in the pGL3 vector. Mutated residues were italicized. MIR376A seed target sequence was underlined. (C and D) Normalized luciferase activity measuremets of lysates from 293T cells that were co-transfected with either MIR-CNT or MIR376A plasmids together with the wild-type or mutant luciferase constructs (mean ± S.D. of independent experiments, n = 3 for ATG4C tests, *p<0,05 and n = 5 for BECN1 tests, ***p<0,01).
Figure 7. Endogenous MIR376A limits starvation-induced autophagy.
(A) Blockage of endogenous MIR376A by Ant-376a, but not CNT-Ant further stimulated starvation (STV)-activated LC3-I to LC3-II conversion in MCF-7 cells. ACTB was used as a loading control. LC3-II/LC3-I densitometric ratios were marked. (B) Ant-376a, but not CNT-Ant resulted in further activation of SQSTM1 protein degradation following starvation in MCF-7 cells. SQSTM1/ACTB ratios were marked.
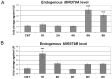
Figure 8. MIR376A and MIR376B level changes in response to starvation stress.
(A and B) TaqMan quantitative PCR (qPCR) analysis of endogenous MIR376A (A) and MIR376B (B) levels under control (CNT, no starvation) or starvation (STV, 1 to 8 hours) conditions. TaqMan qPCR data was normalized using U6 small nuclear 1 (RNU6-1) (U6) mRNA levels (mean ± S.D. of independent experiments, n = 5 for both A and B, ***p<0,01).
Similar articles
- miR-376b controls starvation and mTOR inhibition-related autophagy by targeting ATG4C and BECN1.
Korkmaz G, le Sage C, Tekirdag KA, Agami R, Gozuacik D. Korkmaz G, et al. Autophagy. 2012 Feb 1;8(2):165-76. doi: 10.4161/auto.8.2.18351. Epub 2012 Feb 1. Autophagy. 2012. PMID: 22248718 - Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells.
Zhu H, Wu H, Liu X, Li B, Chen Y, Ren X, Liu CG, Yang JM. Zhu H, et al. Autophagy. 2009 Aug;5(6):816-23. doi: 10.4161/auto.9064. Epub 2009 Aug 20. Autophagy. 2009. PMID: 19535919 Free PMC article. - A fine-tuning mechanism underlying self-control for autophagy: deSUMOylation of BECN1 by SENP3.
Liu K, Guo C, Lao Y, Yang J, Chen F, Zhao Y, Yang Y, Yang J, Yi J. Liu K, et al. Autophagy. 2020 Jun;16(6):975-990. doi: 10.1080/15548627.2019.1647944. Epub 2019 Aug 2. Autophagy. 2020. PMID: 31373534 Free PMC article. - Impaired autophagy and APP processing in Alzheimer's disease: The potential role of Beclin 1 interactome.
Salminen A, Kaarniranta K, Kauppinen A, Ojala J, Haapasalo A, Soininen H, Hiltunen M. Salminen A, et al. Prog Neurobiol. 2013 Jul-Aug;106-107:33-54. doi: 10.1016/j.pneurobio.2013.06.002. Epub 2013 Jul 1. Prog Neurobiol. 2013. PMID: 23827971 Review. - Investigating the role of reactive oxygen species in regulating autophagy.
Gibson SB. Gibson SB. Methods Enzymol. 2013;528:217-35. doi: 10.1016/B978-0-12-405881-1.00013-6. Methods Enzymol. 2013. PMID: 23849868 Review.
Cited by
- Transcriptional and epigenetic regulation of autophagy in aging.
Lapierre LR, Kumsta C, Sandri M, Ballabio A, Hansen M. Lapierre LR, et al. Autophagy. 2015;11(6):867-80. doi: 10.1080/15548627.2015.1034410. Autophagy. 2015. PMID: 25836756 Free PMC article. Review. - Beclin 1 biology and its role in heart disease.
Zhu H, He L. Zhu H, et al. Curr Cardiol Rev. 2015;11(3):229-37. doi: 10.2174/1573403x10666141106104606. Curr Cardiol Rev. 2015. PMID: 25373623 Free PMC article. Review. - MITF-MIR211 axis is a novel autophagy amplifier system during cellular stress.
Ozturk DG, Kocak M, Akcay A, Kinoglu K, Kara E, Buyuk Y, Kazan H, Gozuacik D. Ozturk DG, et al. Autophagy. 2019 Mar;15(3):375-390. doi: 10.1080/15548627.2018.1531197. Epub 2018 Oct 16. Autophagy. 2019. PMID: 30290719 Free PMC article. - miR-378a: a new emerging microRNA in metabolism.
Machado IF, Teodoro JS, Palmeira CM, Rolo AP. Machado IF, et al. Cell Mol Life Sci. 2020 May;77(10):1947-1958. doi: 10.1007/s00018-019-03375-z. Epub 2019 Nov 20. Cell Mol Life Sci. 2020. PMID: 31748917 Free PMC article. Review. - MicroRNA as a Therapeutic Target in Cardiac Remodeling.
Chen C, Ponnusamy M, Liu C, Gao J, Wang K, Li P. Chen C, et al. Biomed Res Int. 2017;2017:1278436. doi: 10.1155/2017/1278436. Epub 2017 Sep 28. Biomed Res Int. 2017. PMID: 29094041 Free PMC article. Review.
References
- Mizushima N (2007) Autophagy: process and function. Genes Dev 21: 2861–2873. - PubMed
- Mizushima N, Yoshimori T, Ohsumi Y (2011) The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 27: 107–132. - PubMed
- Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, et al. (2010) Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev 90: 1383–1435. - PubMed
- Ohsumi Y, Mizushima N (2004) Two ubiquitin-like conjugation systems essential for autophagy. Semin Cell Dev Biol 15: 231–236. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
This work was supported by The Scientific and Technological Research Council of Turkey (TUBITAK) 1001 Grant and Sabanci University. D.G. and A.K are recipients the Turkish Academy of Sciences (TUBA) GEBIP Award. D.G. is a recipient of the EMBO Strategical Development and Installation Grant (EMBO-SDIG) and A.K. is a recipient of the TUBITAK Incentive Award. G.K. and K.A.T. are recipients of Yousef Jameel and TUBITAK-BIDEB PhD Scholarships, respectively. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources