The bacterial cytoplasm has glass-like properties and is fluidized by metabolic activity - PubMed (original) (raw)
The bacterial cytoplasm has glass-like properties and is fluidized by metabolic activity
Bradley R Parry et al. Cell. 2014.
Abstract
The physical nature of the bacterial cytoplasm is poorly understood even though it determines cytoplasmic dynamics and hence cellular physiology and behavior. Through single-particle tracking of protein filaments, plasmids, storage granules, and foreign particles of different sizes, we find that the bacterial cytoplasm displays properties that are characteristic of glass-forming liquids and changes from liquid-like to solid-like in a component size-dependent fashion. As a result, the motion of cytoplasmic components becomes disproportionally constrained with increasing size. Remarkably, cellular metabolism fluidizes the cytoplasm, allowing larger components to escape their local environment and explore larger regions of the cytoplasm. Consequently, cytoplasmic fluidity and dynamics dramatically change as cells shift between metabolically active and dormant states in response to fluctuating environments. Our findings provide insight into bacterial dormancy and have broad implications to our understanding of bacterial physiology, as the glassy behavior of the cytoplasm impacts all intracellular processes involving large components.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
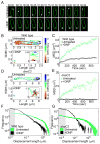
Figure 1. The mobility of crescentin-GFP structures and GFP-LacI-labeled mini-RK2 plasmids is affected by metabolism
(A) Time-lapse montages of crescentin-GFP structures acquired under conditions of growth (M2G) and carbon source depletion. C. crescentus cells (CJW1265) were grown and imaged in M2G, a glucose-based medium (top montage). For carbon starvation, cells were washed into M2 buffer (lacking glucose) and incubated for 3 h before imaging (bottom montage). The scale bar is 1 μm. (B) Two-dimensional trajectories representing 200 min of crescentin-GFP tracking from single C. crescentus cells (CJW1265) under metabolically active (M2G) and metabolically depleted (carbon starvation, late stationary phase, + DNP) conditions. (C) MSD of crescentin-GFP structures in metabolically active (M2G, n = 1,796 trajectories) and energy-depleted (carbon starvation, n = 861 trajectories; stationary phase, n = 718 trajectories; and +DNP, n = 1,943 trajectories) conditions. C. crescentus cells (CJW1265) from late stationary phase cell cultures (OD660 ≥ 1.7) were imaged on agarose pads made with stationary-phase culture supernatant instead of M2G. (D) Two-dimensional trajectories of GFP-LacI-labeled mini-RK2 plasmids overlaid on corresponding phase-contrast images of metabolically active and DNP-treated E. coli cells (JP924). Scale bar is 1 μm. (E) MSD of mini-RK2 plasmids under metabolically active (untreated, n = 497 trajectories) and energy-depleted (+DNP, n = 488 trajectories) conditions. See also Figs. S1–2 and Movies 1–6
Figure 2. GFP-μNS probe dynamics are affected by cellular metabolism
(A) Representative time-lapse montages of GFP-μNS particles (yellow) in E. coli cells (CJW4617) acquired under untreated and DNP-treated conditions (+DNP). The scale bar is 1 μm. Time is min:sec. (B) An example of a two-dimensional trajectory of a GFP-μNS particle in an E. coli cell (CJW4617) with or without DNP treatment. (C) MSD of GFP-μNS particles in metabolically active (untreated, n = 729 trajectories) and DNP-treated (+DNP, n = 643 trajectories) E. coli cells (CJW4617). (D) Two-dimensional trajectory of a GFP-μNS particle in a filamentous E. coli dnaC2 cell (CJW4619) at the restrictive temperature (37ºC) with or without DNP treatment. (E) MSD of GFP-μNS particles in filamentous E. coli dnaC2 cells (CJW4619) at their restrictive temperature with (+DNP, n = 118 trajectories) or without (untreated, n = 192 trajectories) DNP treatment. (F) Histogram of GFP-μNS particle displacements in E. coli cells (CJW4617) with or without DNP treatment. Line width indicates Poisson counting error, and the gray shading delineates the estimated tracking error. Displacements were measured over 15-sec intervals. (G) Histogram of GFP-μNS particle displacements in E. coli dnaC2 cells (CJW4619) at the restrictive temperature (37ºC) with or without DNP treatment. Line width indicates Poisson counting error, and the gray shading delineates the estimated tracking error. Displacements were measured over 15-sec intervals. See also Figs. S2–5 and Movie 7.
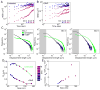
Figure 3. Effect of metabolism on cytoplasmic motion depends on particle size
(A) MSD of GFP-μNS particles of varying binned fluorescence intensities in E. coli cells (CJW4617) under normal growth conditions (M2G). (B) Same as A, but for GFP-μNS particles in E. coli cells (CJW4617) under DNP treatment. (C) Histograms of GFP-μNS particle displacements under untreated and DNP-treated conditions for selected bins. Displacements were measured over 15-sec intervals. Line width indicates Poisson counting error, and the gray shading delineates the estimated tracking error. (D) Mean radius of gyration (Rg) of all trajectories from GFP-μNS particles of a given binned size is plotted as a measure of spatial exploration for untreated and DNP-treated cells. (E) The ratio (RgUntreated-Rg+DNP)/Rg+DNP is plotted as a function of particle size. See also Figs. S6–7.
Figure 4. GFP-μNS particles display non-Gaussian and non-ergodic behavior in the cytoplasm
(A) Experimental distributions of GFP-μNS particle displacements for E. coli cells (CJW4617) under untreated and DNP-treated conditions for a selected size bin (bin 5). Dashed lines are the best fit to a Gaussian distribution. Line width indicates Poisson counting error, and the gray shading delineates the estimated tracking error. Time interval is 15 sec. (B) Non-Gaussian parameter α2 of particle displacement distributions is plotted as a function of particle size for E. coli cells (CJW4617) with or without DNP treatment. (C) Comparison of MSD and MSDτ for selected size bins for cells under untreated conditions. (D) Same as C, but for cells under DNP treatment. (E) The parameter γ is plotted as a function of particle size for cells with or without DNP treatment.
Figure 5. Two subpopulations of GFP-μNS particles exist in both active and inactive cells
(A) Radius of gyration (Rg) of individual trajectories is plotted as a function of GFP-μNS particle size for E. coli cells (CJW4617) under untreated and DNP-treated conditions. The horizontal dashed line delimits slow (Rg<0.3 μm) and fast (_Rg_>0.3 μm) particles. (B) Histogram of Rg for cells under untreated and DNP-treated conditions. The vertical dashed line delimits slow (Rg<0.3 μm) and fast (_Rg_>0.3 μm) particles. The gray shading delineates the estimated tracking error. (C) Fraction of slow particles (Rg<0.3 μm) for trajectories in each size bin for cells with or without DNP treatment. (D) MSD of slow (_Rg_<0.3 μm) and fast (_Rg_>0.3 μm) GFP-μNS particles. (E) Distribution of displacements for slow (Rg<0.3 μm) and fast (_Rg_>0.3 μm) GFP-μNS particles under untreated and DNP-treated conditions. The line width indicates Poisson counting error, and the gray shading delineates the estimated tracking error.
Figure 6. Double-particle tracking and correlation of displacements are consistent with dynamic heterogeneity in the cytoplasm
(A) Example of two-dimensional trajectories of two GFP-μNS particles in a single E. coli cell (CJW4617). The trajectory of the left particle has a radius of gyration Rg = 0.12 μm and the particle has an estimated size d = 144 nm, while Rg = 0.43 μm and d = 158 nm for the right particle. (B) Radius of gyration (Rg) of individual trajectories for pairs of GFP-μNS particles in individual E. coli cells (CJW4617) under metabolically active conditions. Only the results for particle pairs with a difference of particle size of less than 10% are shown. The horizontal dashed line delimits slow (Rg<0.3 μm) and fast (_Rg_>0.3 μm) particles. (C) Plot showing the average displacement length (displacement2) following an initial displacement (displacement1) of a given length for GFP-μNS particles in E. coli cells (CJW4617) under untreated and DNP-treated conditions. The second displacement was signed positive if it was in the same direction as the initial displacement and negative otherwise. Each point represents the average of 700 displacements. Solid lines represent the best fit (displacement2 = − 0.34 displacement1) to the data where displacement1 < 0.2 μm. The time interval is 15 sec.
Comment in
- Cytoplasmic transport: bacteria turn to glass unless kicked.
Janmey PA, MacKintosh FC. Janmey PA, et al. Curr Biol. 2014 Mar 17;24(6):R226-8. doi: 10.1016/j.cub.2014.02.018. Curr Biol. 2014. PMID: 24650906
Similar articles
- Cytoplasmic transport: bacteria turn to glass unless kicked.
Janmey PA, MacKintosh FC. Janmey PA, et al. Curr Biol. 2014 Mar 17;24(6):R226-8. doi: 10.1016/j.cub.2014.02.018. Curr Biol. 2014. PMID: 24650906 - High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics.
Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. Sliusarenko O, et al. Mol Microbiol. 2011 May;80(3):612-27. doi: 10.1111/j.1365-2958.2011.07579.x. Epub 2011 Mar 17. Mol Microbiol. 2011. PMID: 21414037 Free PMC article. - Bend into shape.
de Boer PA. de Boer PA. EMBO J. 2009 May 6;28(9):1193-4. doi: 10.1038/emboj.2009.91. EMBO J. 2009. PMID: 19421162 Free PMC article. No abstract available. - Control of chromosome replication in caulobacter crescentus.
Marczynski GT, Shapiro L. Marczynski GT, et al. Annu Rev Microbiol. 2002;56:625-56. doi: 10.1146/annurev.micro.56.012302.161103. Epub 2002 Jan 30. Annu Rev Microbiol. 2002. PMID: 12142494 Review. - Cell cycle regulation in Caulobacter: location, location, location.
Goley ED, Iniesta AA, Shapiro L. Goley ED, et al. J Cell Sci. 2007 Oct 15;120(Pt 20):3501-7. doi: 10.1242/jcs.005967. J Cell Sci. 2007. PMID: 17928306 Review.
Cited by
- Enzyme activities predicted by metabolite concentrations and solvent capacity in the cell.
Britton S, Alber M, Cannon WR. Britton S, et al. J R Soc Interface. 2020 Oct;17(171):20200656. doi: 10.1098/rsif.2020.0656. Epub 2020 Oct 14. J R Soc Interface. 2020. PMID: 33050777 Free PMC article. - A glass menagerie of low complexity sequences.
Halfmann R. Halfmann R. Curr Opin Struct Biol. 2016 Jun;38:18-25. doi: 10.1016/j.sbi.2016.05.002. Epub 2016 May 31. Curr Opin Struct Biol. 2016. PMID: 27258703 Free PMC article. Review. - Concomitant osmotic and chaotropicity-induced stresses in Aspergillus wentii: compatible solutes determine the biotic window.
de Lima Alves F, Stevenson A, Baxter E, Gillion JL, Hejazi F, Hayes S, Morrison IE, Prior BA, McGenity TJ, Rangel DE, Magan N, Timmis KN, Hallsworth JE. de Lima Alves F, et al. Curr Genet. 2015 Aug;61(3):457-77. doi: 10.1007/s00294-015-0496-8. Epub 2015 Jun 9. Curr Genet. 2015. PMID: 26055444 - RNA polymerase supply and flux through the lac operon in Escherichia coli.
Sendy B, Lee DJ, Busby SJ, Bryant JA. Sendy B, et al. Philos Trans R Soc Lond B Biol Sci. 2016 Nov 5;371(1707):20160080. doi: 10.1098/rstb.2016.0080. Philos Trans R Soc Lond B Biol Sci. 2016. PMID: 27672157 Free PMC article. - Unjamming and emergent nonreciprocity in active ploughing through a compressible viscoelastic fluid.
Banerjee JP, Mandal R, Banerjee DS, Thutupalli S, Rao M. Banerjee JP, et al. Nat Commun. 2022 Aug 4;13(1):4533. doi: 10.1038/s41467-022-31984-z. Nat Commun. 2022. PMID: 35927258 Free PMC article.
References
- Ausmees N, Kuhn JR, Jacobs-Wagner C. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell. 2003;115:705–713. - PubMed
- Berthier L. Dynamic heterogeneity in amorphous materials. Physics. 2011;4:42.
- Brangwynne CP, Koenderink GH, MacKintosh FC, Weitz DA. Intracellular transport by active diffusion. Trends Cell Biol. 2009;19:423–427. - PubMed
Publication types
MeSH terms
Grants and funding
- T32 HG003198/HG/NHGRI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- GM065835/GM/NIGMS NIH HHS/United States
- T32HG 003198/HG/NHGRI NIH HHS/United States
- R01 GM065835/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials