Structure of Clostridium difficile PilJ exhibits unprecedented divergence from known type IV pilins - PubMed (original) (raw)
Structure of Clostridium difficile PilJ exhibits unprecedented divergence from known type IV pilins
Kurt H Piepenbrink et al. J Biol Chem. 2014.
Abstract
Type IV pili are produced by many pathogenic Gram-negative bacteria and are important for processes as diverse as twitching motility, cellular adhesion, and colonization. Recently, there has been an increased appreciation of the ability of Gram-positive species, including Clostridium difficile, to produce Type IV pili. Here we report the first three-dimensional structure of a Gram-positive Type IV pilin, PilJ, demonstrate its incorporation into Type IV pili, and offer insights into how the Type IV pili of C. difficile may assemble and function. PilJ has several unique structural features, including a dual-pilin fold and the incorporation of a structural zinc ion. We show that PilJ is incorporated into Type IV pili in C. difficile and present a model in which the incorporation of PilJ into pili exposes the C-terminal domain of PilJ to create a novel interaction surface.
Keywords: Bacterial Adhesion; Microbiology; Microfilaments; Protein Assembly; Protein Structure.
Figures
FIGURE 1.
PilJ expression and localization. A, electron micrographs stained with anti-PilA1 and anti-PilJ antibodies and immunogold-labeled secondary antibodies with particle sizes 10 nm (PilA1) and 15 nm (PilJ). Nanoparticles associated with pili are labeled with either gray triangles (10 nm, PilA1) or black triangles (15 nm, PilJ). B, quantification of immunogold labeling. The average number of particles per field is shown as a horizontal line. The error bars show one standard deviation.
FIGURE 2.
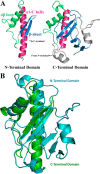
PilJ three-dimensional structure. A, the sequence of PilJ with the secondary structure outlined below. The prepilin leader sequence is shown in black, and the α1-n region is in gray. The N-terminal domain is green, and the C-terminal domain is blue. Helices are shown as boxes, and strands are shown as arrows. B, schematic representation of the structure of PilJ. The N-terminal domain is green, the C-terminal domain is blue, and the zinc atom is magenta. The disordered loop spanning residues 94–102 is indicated with a dotted line. C, the zinc-binding site of PilJ. Cysteines 36, 81. and 111 from the N-terminal domain are in green, histidine 114 from the C-terminal domain is in cyan, and the zinc atom is in magenta. The gray mesh shows the bounds of a 2_Fo_ − Fc electron density map. D, CD spectra of PilJ are shown in increasing concentrations of EDTA.
FIGURE 3.
Structural comparison of the PilJ N-terminal and C-terminal domains. A, the N-terminal and C-terminal domains of PilJ. The initial α helices are in magenta, the αβ loops are in green, and the first two strands of each β-sheet are in blue. B, the N-terminal domain is depicted in green, and the C-terminal domain is in cyan.
FIGURE 4.
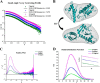
Self-association of Pilin headgroups. A, log-scale intensity plot of SAXS profiles at 2 mg/ml (green), 5 mg/ml (blue), and 10 mg/ml (magenta). B, superimposition of PilJ x-ray crystal structure (cyan ribbon) into envelope calculated for 2 mg/ml PilJ by SASTBX (gray mesh). C, Kratky Plot (I × _q_2 versus q) at 2 mg/ml (green), 5 mg/ml (blue), and 10 mg/ml (magenta). D, radial distribution function calculated by GNOM at 2 mg/ml (green), 5 mg/ml (blue), and 10 mg/ml (magenta). Error bars are shown as hash marks. The inset shows a Gunier plot (ln(I(q)) _versus q_2) of the region used to calculate the radius of gyration.
FIGURE 5.
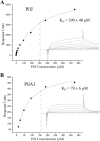
Surface plasmon resonance binding analysis of PilJ and PilA1. Surface plasmon resonance binding of soluble PilJ to PilJ (A) and PilA1 (B) surfaces. The binding titrations are shown as insets, with the steady-state values at each concentration depicted as black squares. The equilibrium fits are shown as gray curves.
FIGURE 6.
Model of PilJ pilus formation. A, model of full-length PilJ. The α1-N helix is shown in gray, the N-terminal domain is in green, and the C-terminal domain is in cyan. B, superimposition of selected regions of PilJ onto TcpA. The PilJ N-terminal domain is shown in green, and the selected portion of the C-terminal domain is in cyan. TcpA is shown in gray. C, space-fill model of truncated PilJ chains modeled into a pilus fragment and superimposed onto electron density from an electron micrograph of the V. cholera TCP (gray mesh). Each chain of the truncated PilJ model is colored individually. D, space-fill and ribbon model of a PilJ pilus formed from full-length PilJ; each chain is colored individually.
FIGURE 7.
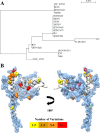
Sequence variation in PilJ. A, unrooted phylogenic tree of C. difficile strains by PilJ sequence. B, model of full-length PilJ. Polymorphic residues in PilJ are represented with side chains rendered as spheres, colored by the number of variations from the reference sequence within the set of 20 C. difficile strains compared here. The blue surface shows the area of each pilin occluded from solvent by the formation of a pilus fragment and hence potentially part of a pilin-pilin interface.
Similar articles
- Structural and evolutionary analyses show unique stabilization strategies in the type IV pili of Clostridium difficile.
Piepenbrink KH, Maldarelli GA, Martinez de la Peña CF, Dingle TC, Mulvey GL, Lee A, von Rosenvinge E, Armstrong GD, Donnenberg MS, Sundberg EJ. Piepenbrink KH, et al. Structure. 2015 Feb 3;23(2):385-96. doi: 10.1016/j.str.2014.11.018. Epub 2015 Jan 15. Structure. 2015. PMID: 25599642 Free PMC article. - Structural and functional studies of the Pseudomonas aeruginosa minor pilin, PilE.
Nguyen Y, Harvey H, Sugiman-Marangos S, Bell SD, Buensuceso RN, Junop MS, Burrows LL. Nguyen Y, et al. J Biol Chem. 2015 Oct 30;290(44):26856-65. doi: 10.1074/jbc.M115.683334. Epub 2015 Sep 10. J Biol Chem. 2015. PMID: 26359492 Free PMC article. - Type IV pili are involved in phenotypes associated with Clostridioides difficile pathogenesis.
Ouyang Z, Zhao H, Zhao M, Yang Y, Zhao J. Ouyang Z, et al. Crit Rev Microbiol. 2024 Nov;50(6):1011-1019. doi: 10.1080/1040841X.2023.2235002. Epub 2023 Jul 15. Crit Rev Microbiol. 2024. PMID: 37452617 Review. - Recognition of extracellular DNA by type IV pili promotes biofilm formation by Clostridioides difficile.
Ronish LA, Sidner B, Yu Y, Piepenbrink KH. Ronish LA, et al. J Biol Chem. 2022 Oct;298(10):102449. doi: 10.1016/j.jbc.2022.102449. Epub 2022 Sep 3. J Biol Chem. 2022. PMID: 36064001 Free PMC article. - Pilins in gram-positive bacteria: A structural perspective.
Krishnan V. Krishnan V. IUBMB Life. 2015 Jul;67(7):533-43. doi: 10.1002/iub.1400. Epub 2015 Jul 14. IUBMB Life. 2015. PMID: 26178080 Review.
Cited by
- Identification, immunogenicity, and cross-reactivity of type IV pilin and pilin-like proteins from Clostridium difficile.
Maldarelli GA, De Masi L, von Rosenvinge EC, Carter M, Donnenberg MS. Maldarelli GA, et al. Pathog Dis. 2014 Aug;71(3):302-14. doi: 10.1111/2049-632X.12137. Epub 2014 Feb 18. Pathog Dis. 2014. PMID: 24550179 Free PMC article. - Crystal Structure of the Minor Pilin CofB, the Initiator of CFA/III Pilus Assembly in Enterotoxigenic Escherichia coli.
Kolappan S, Ng D, Yang G, Harn T, Craig L. Kolappan S, et al. J Biol Chem. 2015 Oct 23;290(43):25805-18. doi: 10.1074/jbc.M115.676106. Epub 2015 Aug 31. J Biol Chem. 2015. PMID: 26324721 Free PMC article. - Type IV pili promote early biofilm formation by Clostridium difficile.
Maldarelli GA, Piepenbrink KH, Scott AJ, Freiberg JA, Song Y, Achermann Y, Ernst RK, Shirtliff ME, Sundberg EJ, Donnenberg MS, von Rosenvinge EC. Maldarelli GA, et al. Pathog Dis. 2016 Aug;74(6):ftw061. doi: 10.1093/femspd/ftw061. Epub 2016 Jun 30. Pathog Dis. 2016. PMID: 27369898 Free PMC article. - Some of the most interesting CASP11 targets through the eyes of their authors.
Kryshtafovych A, Moult J, Baslé A, Burgin A, Craig TK, Edwards RA, Fass D, Hartmann MD, Korycinski M, Lewis RJ, Lorimer D, Lupas AN, Newman J, Peat TS, Piepenbrink KH, Prahlad J, van Raaij MJ, Rohwer F, Segall AM, Seguritan V, Sundberg EJ, Singh AK, Wilson MA, Schwede T. Kryshtafovych A, et al. Proteins. 2016 Sep;84 Suppl 1(Suppl Suppl 1):34-50. doi: 10.1002/prot.24942. Epub 2015 Nov 16. Proteins. 2016. PMID: 26473983 Free PMC article. - Cyclic di-GMP riboswitch-regulated type IV pili contribute to aggregation of Clostridium difficile.
Bordeleau E, Purcell EB, Lafontaine DA, Fortier LC, Tamayo R, Burrus V. Bordeleau E, et al. J Bacteriol. 2015 Mar;197(5):819-32. doi: 10.1128/JB.02340-14. Epub 2014 Dec 15. J Bacteriol. 2015. PMID: 25512308 Free PMC article.
References
- Strom M. S., Lory S. (1993) Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol 47, 565–596 - PubMed
- Girón J. A., Ho A. S., Schoolnik G. K. (1991) An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science 254, 710–713 - PubMed
- Lee K. K., Sheth H. B., Wong W. Y., Sherburne R., Paranchych W., Hodges R. S., Lingwood C. A., Krivan H., Irvin R. T. (1994) The binding of Pseudomonas aeruginosa pili to glycosphingolipids is a tip-associated event involving the C-terminal region of the structural pilin subunit. Mol. Microbiol. 11, 705–713 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI114902/AI/NIAID NIH HHS/United States
- T32 AI095190/AI/NIAID NIH HHS/United States
- T32 DK067872/DK/NIDDK NIH HHS/United States
- F32 AI110045/AI/NIAID NIH HHS/United States
- R21 AI105881/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources