A genetic screen identifies an LKB1-MARK signalling axis controlling the Hippo-YAP pathway - PubMed (original) (raw)
doi: 10.1038/ncb2884. Epub 2013 Dec 22.
Jianlong Sun 1, Allison Lau 2, Stephen Curtis 3, Jeffrey Goldsmith 4, Victor L Fox 5, Chongjuan Wei 6, Marsha Frazier 6, Owen Samson 7, Kwok-Kin Wong, Carla Kim 3, Fernando D Camargo 1
Affiliations
- PMID: 24362629
- PMCID: PMC4159053
- DOI: 10.1038/ncb2884
A genetic screen identifies an LKB1-MARK signalling axis controlling the Hippo-YAP pathway
Morvarid Mohseni et al. Nat Cell Biol. 2014 Jan.
Erratum in
- Nat Cell Biol. 2014 Feb;16(2):200. Wong, Kwok-Kim [corrected to Wong, Kwok-Kin]
Abstract
The Hippo-YAP pathway is an emerging signalling cascade involved in the regulation of stem cell activity and organ size. To identify components of this pathway, we performed an RNAi-based kinome screen in human cells. Our screen identified several kinases not previously associated with Hippo signalling that control multiple cellular processes. One of the hits, LKB1, is a common tumour suppressor whose mechanism of action is only partially understood. We demonstrate that LKB1 acts through its substrates of the microtubule affinity-regulating kinase family to regulate the localization of the polarity determinant Scribble and the activity of the core Hippo kinases. Our data also indicate that YAP is functionally important for the tumour suppressive effects of LKB1. Our results identify a signalling axis that links YAP activation with LKB1 mutations, and have implications for the treatment of LKB1-mutant human malignancies. In addition, our findings provide insight into upstream signals of the Hippo-YAP signalling cascade.
Figures
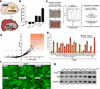
Figure 1. Kinome RNAi screen identifies novel regulators of Hippo/YAP signaling
A) Graphical representation of Yap/Taz mediated STBS reporter activation in vitro. B) Validation of STBS reporter sensitivity using siRNA knockdown of known components of Hippo signaling. CTR=Scrambled negative control. C) Schematic of RNAi screening strategy. The RNAi screen was performed in 96 well plates using a stably expressing 293T-STBS-mCherry reporter cell line. Activation of the STBS-mCherry reporter was visualized 4 days following siRNA transfection. Fluorescence intensity was captured by flow cytometry. Statistical analysis was performed to identify genes for secondary screening and final selection of hits. D) Mean Z-score and mCherry reporter fold change (vs. scrambled controls) values for each triplicate siRNA oligo were plotted to identify hits that statistical thresholds of Z-score >2 and fold change greater than 4. Highlighted box represents hits satisfying these thresholds. Green filled circles represent siRNA knockdown of LATS2 as a positive control. E) A secondary siRNA screen identifies kinases that reproducibly raise STBS-mcherry reporter activity, performed using an alternative siRNA oligo source using two reporter systems. F) YAP immunolocalization in HaCaT cells following siRNA knockdown of kinases that regulate STBS reporter activity. G) Immunoblot for S127 Yap phosphorylation following siRNA knockdown of kinases from secondary screen. CTR represents Scr siRNA and NF2 siRNA is used as a positive control. Error bars represent ± SD from triplicate samples. Scale bars, 200 µm
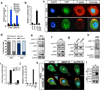
Figure 2. LKB1 regulates YAP activity through the Hippo kinases
A, B) LKB1 knockdown induces YAP target gene expression of AMOTL2and Cyr61 and TEAD reporter activation and is dependent on Yap expression. C) Immunofluorescence for F-actin (red), Yap (green) and nuclei (blue) in LS174T (W4) cells. Dox-inducible LKB1 activation after 24 hours results in LKB1-dependent cell polarization and Yap nuclear to cytoplasmic translocation. D) Quantification of cell polarization and Yap subcellular localization following Dox administration. E) MST1 activity in W4 cells is induced upon LKB1 activation (+Dox). Note increased MST1/2 phosphorylation in the full length and cleaved forms of MST1 and increase in levels of cleaved active MST1 peptide. F) Activity of Lats1/2 is increased upon LKB1 activation as measured by phosphorylation at Thr1079 LATS1/2. G) LATS1/2 phosphorylation at Thr1079 is abolished upon siRNA knockdown of LKB1 in MCF7 cells. H, I) Western blot analysis and qPCR of Yap target gene performed on liver lysates derived from Adeno-cre infected Lkb1 +/+ or Lkb1f/f mice 3months post infection. Both lines of mice also carried a p53 homozygous floxed allele. LKB1 deficiency leads to an overall decrease in cleaved activated MST1 and Thr183/Thr180 MST1/2 phosphorylation and increase in CTGF and CYR61 expression. J) Overexpression of LATS1, LATS2 and MOB1 in LKB1 knockdown 293T cells can restore STBS reporter activity. K) Knockdown of MST1/2 and LATS1/2 in Dox treated W4 cells suppresses LKB1-driven cytoplasmic translocation of YAP (green) when compared to the scrambled negative control (siCTR). L) Endogenous co-immunoprecipitation experiments using 293T cells expressing demonstrate physical association of Lkb1 with LATS1 and MST1. Error bars represent ± SD from triplicate samples. **, P≤0.01. Scale bars, 20 µm
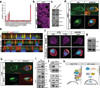
Figure 3. MARKs/SCRIB act downstream of LKB1 to regulate Hippo/YAP
A) Small scale RNAi screen on downstream substrates of LKB1 in 293T STBS-luc cells. Error bars represent ± SD from triplicate samples. B, C) Nuclear Yap accumulation and decreases in Lats and Yap phosphorylation following knockdown of MARKs. D) Suppression of LKB1-driven cytoplasmic translocation of YAP following MARK4 knockdown in Dox-treated W4 cells. Scale bars, 20 µm. E) Confocal immunofluorescent and Z-stack analysis for Scribble (SCRIB, green), F-actin (red) and nuclei (blue) in LKB1 and MARKs knockdown in MCF7 cells. Note mislocalization of SCRIB following LKB1 or MARKs silencing. F) Immunofluorescence in W4 cells demonstrates that LKB1 activation leads to SCRIB re-localization to the cell membrane and actin cap and require MARK expression. G) Knockdown of SCRIB in 293T cells reduces S127 Yap phosphorylation. H) Knockdown of SCRIB in Dox-induced LKB1 activated W4 cells suppressed YAP re-localization to the cytoplasm/actin cap. Scale bars, 20 µm. I) Endogenous co-immunoprecipitation of MARK1 demonstrates biochemical interactions with LKB1, SCRIB, MST1 and LATS1. J) Immunoblot for MST1 and LATS1, in SCRIB immunopreciptates derived from 293T cells with concomitant knockdown of MARK1. Adjusted lysate amounts were used for the IP to obtain equal levels of immunoprecipitated SCRIB. K) Diagram showing LKB1/MARK signaling axis controlling SCRIB localization and MST/LATS activation.
Figure 4. YAP activity in LKB1 deficient tumors
A) Immunohistochemistry for YAP on Ad-cre administered KrasG12D mutant (K) and KrasG12D/LKB1fl/fl (KL) on Grade I, II lung adenocarcinoma. Representative picture shown, n = 5 for each genotype. B) Gene set enrichment using a Hippo signature on K versus KL murine lung tumors. C) Western blot analysis for activated, phosphorylated and cleaved, MST1/2 and S127 Yap phosphorylation on K and KL murine lung nodules (n=3 mice). D) Immunohistochemistry for Scribble localization in K and KL lung adenocarcinomas, (n=5 mice). E) YAP localization assessed by immunohistochemistry in human intestinal tissue and Peutz-Jeghers (PJ) intestinal polyps. Representative data, n=3. F) Immunohistochemistry for YAP localization and expression in normal ductal tissue compared with ductal breast adenocarcinoma, and in normal human liver compared with metastatic liver adenocarcinoma derived from a Peutz-Jeghers syndrome patient (G). H) Subcutaneous xenograft assay using W4 cells, and W4 cells that co-express doxycycline inducible YapS127A. Tumor volumes for non-induced and induced tumors are shown. Representative tumors from non-induced and induced W4 and W4TetOYap xenografts are displayed. I-J) Genetic deletion of LKB1 and YAP1 in the liver following Ad-cre display restoration of organ size and hepatocyte proliferation. Animals received Ad-Cre at 1 month of age and tissues were collected 2.5 months later. Error bars represent ± SD from n=5 mice.
Similar articles
- Hippo component YAP promotes focal adhesion and tumour aggressiveness via transcriptionally activating THBS1/FAK signalling in breast cancer.
Shen J, Cao B, Wang Y, Ma C, Zeng Z, Liu L, Li X, Tao D, Gong J, Xie D. Shen J, et al. J Exp Clin Cancer Res. 2018 Jul 28;37(1):175. doi: 10.1186/s13046-018-0850-z. J Exp Clin Cancer Res. 2018. PMID: 30055645 Free PMC article. - Regulation of the Hippo-YAP Pathway by Glucose Sensor O-GlcNAcylation.
Peng C, Zhu Y, Zhang W, Liao Q, Chen Y, Zhao X, Guo Q, Shen P, Zhen B, Qian X, Yang D, Zhang JS, Xiao D, Qin W, Pei H. Peng C, et al. Mol Cell. 2017 Nov 2;68(3):591-604.e5. doi: 10.1016/j.molcel.2017.10.010. Mol Cell. 2017. PMID: 29100056 - PAR1 participates in the ability of multidrug resistance and tumorigenesis by controlling Hippo-YAP pathway.
Fujimoto D, Ueda Y, Hirono Y, Goi T, Yamaguchi A. Fujimoto D, et al. Oncotarget. 2015 Oct 27;6(33):34788-99. doi: 10.18632/oncotarget.5858. Oncotarget. 2015. PMID: 26431277 Free PMC article. - Hippo-YAP signaling pathway: A new paradigm for cancer therapy.
Ma Y, Yang Y, Wang F, Wei Q, Qin H. Ma Y, et al. Int J Cancer. 2015 Nov 15;137(10):2275-86. doi: 10.1002/ijc.29073. Epub 2014 Jul 22. Int J Cancer. 2015. PMID: 25042563 Review. - Control of Proliferation and Cancer Growth by the Hippo Signaling Pathway.
Ehmer U, Sage J. Ehmer U, et al. Mol Cancer Res. 2016 Feb;14(2):127-40. doi: 10.1158/1541-7786.MCR-15-0305. Epub 2015 Oct 2. Mol Cancer Res. 2016. PMID: 26432795 Free PMC article. Review.
Cited by
- New Insights into YAP/TAZ-TEAD-Mediated Gene Regulation and Biological Processes in Cancer.
Zhao Y, Sheldon M, Sun Y, Ma L. Zhao Y, et al. Cancers (Basel). 2023 Nov 21;15(23):5497. doi: 10.3390/cancers15235497. Cancers (Basel). 2023. PMID: 38067201 Free PMC article. Review. - ZDHHC7-mediated S-palmitoylation of Scribble regulates cell polarity.
Chen B, Zheng B, DeRan M, Jarugumilli GK, Fu J, Brooks YS, Wu X. Chen B, et al. Nat Chem Biol. 2016 Sep;12(9):686-93. doi: 10.1038/nchembio.2119. Epub 2016 Jul 4. Nat Chem Biol. 2016. PMID: 27380321 Free PMC article. - Clinical potential of the Hippo-YAP pathway in bladder cancer.
Cheng X, Lou K, Ding L, Zou X, Huang R, Xu G, Zou J, Zhang G. Cheng X, et al. Front Oncol. 2022 Jul 15;12:925278. doi: 10.3389/fonc.2022.925278. eCollection 2022. Front Oncol. 2022. PMID: 35912245 Free PMC article. Review. - LKB1 and YAP phosphorylation play important roles in Celastrol-induced β-catenin degradation in colorectal cancer.
Wang S, Ma K, Zhou C, Wang Y, Hu G, Chen L, Li Z, Hu C, Xu Q, Zhu H, Liu M, Xu N. Wang S, et al. Ther Adv Med Oncol. 2019 Apr 22;11:1758835919843736. doi: 10.1177/1758835919843736. eCollection 2019. Ther Adv Med Oncol. 2019. PMID: 31040884 Free PMC article. - CDC42 controlled apical-basal polarity regulates intestinal stem cell to transit amplifying cell fate transition via YAP-EGF-mTOR signaling.
Zhang Z, Zhang F, Davis AK, Xin M, Walz G, Tian W, Zheng Y. Zhang Z, et al. Cell Rep. 2022 Jan 11;38(2):110009. doi: 10.1016/j.celrep.2021.110009. Cell Rep. 2022. PMID: 35021092 Free PMC article.
References
- Takebe N, Harris PJ, Warren RQ, Ivy SP. Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat Rev Clin Oncol. 2011;8:97–106. - PubMed
- Ota M, Sasaki H. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development. 2008;135:4059–4069. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases