Chromatin modifiers and remodellers: regulators of cellular differentiation - PubMed (original) (raw)
Review
Chromatin modifiers and remodellers: regulators of cellular differentiation
Taiping Chen et al. Nat Rev Genet. 2014 Feb.
Abstract
Cellular differentiation is, by definition, epigenetic. Genome-wide profiling of pluripotent cells and differentiated cells suggests global chromatin remodelling during differentiation, which results in a progressive transition from a fairly open chromatin configuration to a more compact state. Genetic studies in mouse models show major roles for a variety of histone modifiers and chromatin remodellers in key developmental transitions, such as the segregation of embryonic and extra-embryonic lineages in blastocyst stage embryos, the formation of the three germ layers during gastrulation and the differentiation of adult stem cells. Furthermore, rather than merely stabilizing the gene expression changes that are driven by developmental transcription factors, there is emerging evidence that chromatin regulators have multifaceted roles in cell fate decisions.
Figures
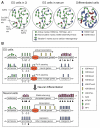
Figure 1. Chromatin states in pluripotent and differentiated cells
(A) ES cells have a globally `open' chromatin state, characterized by the enrichment of active histone marks such as histone acetylation and H3K4 methylation, whereas differentiated cells have a more compact chromatin state, characterized by expanded domains of repressive histone marks such as H3K27me3, H3K9me2 and H3K9me3. ES cells cultured in 2i medium are highly similar to the naïve pluripotent cells in ICM of blastocysts, and ES cells cultured in serum are more heterogeneous. Some H3K4me3 and H3K27me3 `bivalent' marks may reflect the cellular heterogeneity, especially when ES cells are cultured in serum. (B) Major chromatin features in different genomic regions. In ES cells, the enhancers of both pluripotency-associated genes and developmental genes are enriched with H3K4me1 and p300. The presence of H3K27ac makes enhancers of pluripotency-associated genes active, whereas the lack of H3K27ac and enrichment of H3K27me3 keep enhancers of developmental lineage-commitment genes in a `poised' state. The promoters of pluripotency-associated genes and lineage-commitment genes are also believed to be active and `poised', respectively. Transcriptional elongation is prevented at lineage-commitment genes due to promoter-proximal RNA polymerase II (Pol II) pausing. Upon differentiation toward a specific lineage (e.g. neural lineage), lineage-specific genes acquire active marks at enhancer and promoter regions, and Pol II pausing is released to allow productive elongation. Genes of other lineages lose enhancer marks and gain H3K27me3 at promoters, resulting in repression. Pluripotency-associated genes gain H3K9 methylation and DNA methylation and become stably silenced. During differentiation, heterochromatin regions — characterized by H3K9me2 and H3K9me3, HP1 binding and DNA methylation — are expanded and become more condensed. H3K27me3 in intergenic regions and repressed genes also expands to large domains.
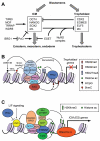
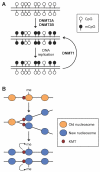
Figure 2. Chromatin regulators involved in the segregation of embryonic and extraembryonic lineages during preimplantation development
(A) The transcription factors specifying the inner cell mass (ICM) and trophoblast cells reciprocally antagonize each other. Chromatin regulators maintain the identity of ICM cells by repressing the trophectoderm transcriptional program, preventing differentiation toward the three germ layers, promoting the expression of pluripotency factors, and functioning as co-regulators or effectors of pluripotency factors. Chromatin regulators that promote trophoblast differentiation are less well understood. (B) In ICM cells, ESET functions as an OCT4 co-repressor to repress trophoblast genes by depositing H3K9me2 and H3K9me3. The NuRD complex is also involved in the repression of trophoblast genes, possibly by multiple mechanisms. The NuRD complex has chromatin-remodeling activity that alters DNA–histone interactions. HDAC1 and HDAC2, which are components of NuRD, deacetylate histones, and deacetylation of H3K27 has been shown to facilitate PRC2 binding and H3K27 methylation. NuRD may also promote DNA methylation by inducing DNMT3B expression. Recent evidence suggests that the MBD3 subunit can bind 5-hydroxymethylcytosine (5hmC). (C) In ICM cells, pluripotency factors, such as OCT4, NANOG and SOX2, recruit histone-modifying enzymes (e.g. TIP60, MOF and MLL complexes) and chromatin-remodeling complexes (e.g. SWI/SNF-BRG1) to ICM-specifying genes (including Oct4, Nanog and Sox2 themselves) and their targets to create `open' chromatin states. One of the effects of the SWI/SNF-BRG1 complex is to maintain chromatin accessibility at STAT3-binding targets by opposing PcG-mediated repression, thus enhancing LIF signaling.
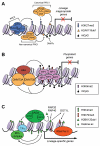
Figure 3. Chromatin regulators involved in gene regulation during postimplantation development and cellular differentiation
(A) PcG proteins play important roles in repressing developmental genes. The PRC2 complex deposits the repressive H3K27me3 marks, which create binding sites for the canonical, CBX-containing PRC1 complex. The noncanonical, RYBP-containing PRC1 complex binds to chromation via H3K27me3-independent mechanisms. RING1A and RING1B, components of the PRC1 complexes, mediate the repressive H2AK119ub1 modification. DNA methylation, mediated by DNA methyltransferases (DNMTs), is also important in repressing lineage-commitment genes, especially those with low CpG-content promoters. (B) G9A is critical for silencing pluripotency-associated genes in postimplantation embryos and differentiated cells. G9A mediates H3K9me2, which induces heterochromatinization by recruiting HP1. G9A also recruits DNMT3A and DNMT3B, which initiate de novo DNA methylation. (C) In differentiating cells, lineage-specific transcription factors (TFs) recruit chromatin-modifying complexes, such as the MLL complex and the SAGA complex, to lineage-specific genes to create `open' chromatin states. The MLL complex deposits the active H3K4me3 marks. The SAGA complex has at least two enzymatic activities: histone acetylation by GCN5 and H2BK120 deubiquitylation by USP22. H2BK120ub1, mediated by RNF20 and RNF40, is preferentially enriched in the coding regions of differentiation-related genes, but not pluripotency-associated genes. H2BK120ub1 has been shown to promote MLL-mediated H3K4 methylation and DOT1L-mediated H3K79 methylation.
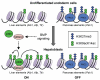
Figure 4. Epigenetic pre-patterning for lineage specification
In multipotent endoderm cells, regulatory elements of liver-specific genes and pancreas-specific genes are `pre-patterned' with distinct chromatin marks; both the active H3K9acK14ac marks and the repressive H3K27me3 mark are enriched in the regulatory elements of pancreas-expressed genes (such as Pdx1), but they are low or undetectable in the regulatory elements of liver-expressed genes (such as Alb1, Afp and Ttr). In response to BMP signaling, SMAD4 recruits p300 to the regulatory elements of liver-expressed genes to stimulate histone acetylation and induce hepatic specification. Meanwhile, the PRC2 complex maintains the level of H3K27me3 in the regulatory elements of pancreas-expressed genes to prevent pancreas specification.
Figure 5. Inheritance of DNA methylation and histone methylation marks through DNA replication
(A) Semi-conservative maintenance of symmetric CpG methylation. During early embryogenesis, DNA methylation patterns are established by the de novo DNA methyltransferases DNMT3A and DNMT3B. After each round of DNA replication, the maintenance methyltransferase DNMT1 `copies' the CpG methylation patterns from the parental strand onto the daughter strand. (B) Role of lysine methyltransferases (KMTs) in maintaining histone methylation. During DNA replication, methylated histones are replaced by unmodified histones, but KMTs remain associated with newly replicated DNA at specific loci. Following DNA replication, the enzymes methylate the newly incorporated histones to re-establish the methylation patterns.
Similar articles
- Dynamic expression of chromatin modifiers during developmental transitions in mouse preimplantation embryos.
Nestorov P, Hotz HR, Liu Z, Peters AH. Nestorov P, et al. Sci Rep. 2015 Sep 25;5:14347. doi: 10.1038/srep14347. Sci Rep. 2015. PMID: 26403153 Free PMC article. - Emerging roles of the histone chaperone CAF-1 in cellular plasticity.
Cheloufi S, Hochedlinger K. Cheloufi S, et al. Curr Opin Genet Dev. 2017 Oct;46:83-94. doi: 10.1016/j.gde.2017.06.004. Epub 2017 Jul 7. Curr Opin Genet Dev. 2017. PMID: 28692904 Free PMC article. Review. - Dynamic changes of the epigenetic landscape during cellular differentiation.
Dambacher S, de Almeida GP, Schotta G. Dambacher S, et al. Epigenomics. 2013 Dec;5(6):701-13. doi: 10.2217/epi.13.67. Epigenomics. 2013. PMID: 24283883 Review. - Multi-omics profiling of mouse gastrulation at single-cell resolution.
Argelaguet R, Clark SJ, Mohammed H, Stapel LC, Krueger C, Kapourani CA, Imaz-Rosshandler I, Lohoff T, Xiang Y, Hanna CW, Smallwood S, Ibarra-Soria X, Buettner F, Sanguinetti G, Xie W, Krueger F, Göttgens B, Rugg-Gunn PJ, Kelsey G, Dean W, Nichols J, Stegle O, Marioni JC, Reik W. Argelaguet R, et al. Nature. 2019 Dec;576(7787):487-491. doi: 10.1038/s41586-019-1825-8. Epub 2019 Dec 11. Nature. 2019. PMID: 31827285 Free PMC article. - Transcription factor binding dynamics during human ES cell differentiation.
Tsankov AM, Gu H, Akopian V, Ziller MJ, Donaghey J, Amit I, Gnirke A, Meissner A. Tsankov AM, et al. Nature. 2015 Feb 19;518(7539):344-9. doi: 10.1038/nature14233. Nature. 2015. PMID: 25693565 Free PMC article.
Cited by
- Active enhancers: recent research advances and insights into disease.
Zhang J, Wang Q, Liu J, Duan Y, Liu Z, Zhang Z, Li C. Zhang J, et al. Biol Direct. 2024 Nov 12;19(1):112. doi: 10.1186/s13062-024-00559-x. Biol Direct. 2024. PMID: 39533395 Free PMC article. Review. - Chromatin remodeller Chd7 is developmentally regulated in the neural crest by tissue-specific transcription factors.
Williams RM, Taylor G, Ling ITC, Candido-Ferreira I, Fountain DM, Mayes S, Ateş-Kalkan PS, Haug JO, Price AJ, McKinney SA, Bozhilovh YK, Tyser RCV, Srinivas S, Hughes JR, Sauka-Spengler T. Williams RM, et al. PLoS Biol. 2024 Oct 17;22(10):e3002786. doi: 10.1371/journal.pbio.3002786. eCollection 2024 Oct. PLoS Biol. 2024. PMID: 39418292 Free PMC article. - An RNA-centric view of transcription and genome organization.
Henninger JE, Young RA. Henninger JE, et al. Mol Cell. 2024 Oct 3;84(19):3627-3643. doi: 10.1016/j.molcel.2024.08.021. Mol Cell. 2024. PMID: 39366351 Review. - Mucosa-like differentiation of head and neck cancer cells is inducible and drives the epigenetic loss of cell malignancy.
Oppel F, Gendreizig S, Martinez-Ruiz L, Florido J, López-Rodríguez A, Pabla H, Loganathan L, Hose L, Kühnel P, Schmidt P, Schürmann M, Neumann JM, Viyof Ful F, Scholtz LU, Ligum D, Brasch F, Niehaus K, Escames G, Busche T, Kalinowski J, Goon P, Sudhoff H. Oppel F, et al. Cell Death Dis. 2024 Oct 2;15(10):724. doi: 10.1038/s41419-024-07065-y. Cell Death Dis. 2024. PMID: 39358322 Free PMC article. - WDR77 in Pan-Cancer: Revealing expression patterns, genetic insights, and functional roles across diverse tumor types, with a spotlight on colorectal cancer.
Wang Y, Wu Q, Liu J, Wang X, Xie J, Fu X, Li Y. Wang Y, et al. Transl Oncol. 2024 Nov;49:102089. doi: 10.1016/j.tranon.2024.102089. Epub 2024 Aug 24. Transl Oncol. 2024. PMID: 39182364 Free PMC article.
References
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nature Rev. Genet. 2013;14:204–20. - PubMed
- Skene PJ, Henikoff S. Histone variants in pluripotency and disease. Development. 2013;140:2513–24. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA016672/CA/NCI NIH HHS/United States
- GM096472/GM/NIGMS NIH HHS/United States
- R01 GM067718/GM/NIGMS NIH HHS/United States
- R01 GM096472/GM/NIGMS NIH HHS/United States
- GM067718/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources