Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer - PubMed (original) (raw)
Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer
Balázs Győrffy et al. PLoS One. 2013.
Erratum in
- PLoS One. 2014;9(10):e111842
Abstract
In the last decade, optimized treatment for non-small cell lung cancer had lead to improved prognosis, but the overall survival is still very short. To further understand the molecular basis of the disease we have to identify biomarkers related to survival. Here we present the development of an online tool suitable for the real-time meta-analysis of published lung cancer microarray datasets to identify biomarkers related to survival. We searched the caBIG, GEO and TCGA repositories to identify samples with published gene expression data and survival information. Univariate and multivariate Cox regression analysis, Kaplan-Meier survival plot with hazard ratio and logrank P value are calculated and plotted in R. The complete analysis tool can be accessed online at: www.kmplot.com/lung. All together 1,715 samples of ten independent datasets were integrated into the system. As a demonstration, we used the tool to validate 21 previously published survival associated biomarkers. Of these, survival was best predicted by CDK1 (p<1E-16), CD24 (p<1E-16) and CADM1 (p = 7E-12) in adenocarcinomas and by CCNE1 (p = 2.3E-09) and VEGF (p = 3.3E-10) in all NSCLC patients. Additional genes significantly correlated to survival include RAD51, CDKN2A, OPN, EZH2, ANXA3, ADAM28 and ERCC1. In summary, we established an integrated database and an online tool capable of uni- and multivariate analysis for in silico validation of new biomarker candidates in non-small cell lung cancer.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
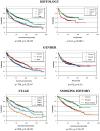
Figure 1. Survival characteristics of the patients included in the database including histology of adenocarcinoma (adeno), squamous cell carcinoma (SCC) and large cell carcinoma (large), gender, stage (only with overall survival) and smoking history.
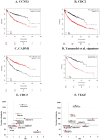
Figure 2. Validation of 29 previously published NSCLC biomarkers.
Meta-analysis of these genes and signatures in the respective sample cohort yielded CCNE1, CDC2 and CADM1 as the best performing individual genes (A–C) and the signature of Yamauchi et al. (D). A funnel plot depicting the hazard ratios (with confidence intervals) versus sample number for CDC2 and VEGF shows more reliable estimation with larger database sizes (E–F).
Similar articles
- A robust six-gene prognostic signature for prediction of both disease-free and overall survival in non-small cell lung cancer.
Zuo S, Wei M, Zhang H, Chen A, Wu J, Wei J, Dong J. Zuo S, et al. J Transl Med. 2019 May 14;17(1):152. doi: 10.1186/s12967-019-1899-y. J Transl Med. 2019. PMID: 31088477 Free PMC article. - Identification and validation of key genes with prognostic value in non-small-cell lung cancer via integrated bioinformatics analysis.
Wang L, Qu J, Liang Y, Zhao D, Rehman FU, Qin K, Zhang X. Wang L, et al. Thorac Cancer. 2020 Apr;11(4):851-866. doi: 10.1111/1759-7714.13298. Epub 2020 Feb 14. Thorac Cancer. 2020. PMID: 32059076 Free PMC article. - miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients.
Lánczky A, Nagy Á, Bottai G, Munkácsy G, Szabó A, Santarpia L, Győrffy B. Lánczky A, et al. Breast Cancer Res Treat. 2016 Dec;160(3):439-446. doi: 10.1007/s10549-016-4013-7. Epub 2016 Oct 15. Breast Cancer Res Treat. 2016. PMID: 27744485 - Comprehensive evaluation of published gene expression prognostic signatures for biomarker-based lung cancer clinical studies.
Tang H, Wang S, Xiao G, Schiller J, Papadimitrakopoulou V, Minna J, Wistuba II, Xie Y. Tang H, et al. Ann Oncol. 2017 Apr 1;28(4):733-740. doi: 10.1093/annonc/mdw683. Ann Oncol. 2017. PMID: 28200038 Free PMC article. Review. - The prognostic value of long non coding RNAs in non small cell lung cancer: A meta-analysis.
Wang M, Ma X, Zhu C, Guo L, Li Q, Liu M, Zhang J. Wang M, et al. Oncotarget. 2016 Dec 6;7(49):81292-81304. doi: 10.18632/oncotarget.13223. Oncotarget. 2016. PMID: 27833074 Free PMC article. Review.
Cited by
- TMEM52B suppression promotes cancer cell survival and invasion through modulating E-cadherin stability and EGFR activity.
Lee Y, Ko D, Yoon J, Lee Y, Kim S. Lee Y, et al. J Exp Clin Cancer Res. 2021 Mar 1;40(1):58. doi: 10.1186/s13046-021-01828-7. J Exp Clin Cancer Res. 2021. PMID: 33641663 Free PMC article. - The vascular nature of lung-resident mesenchymal stem cells.
Steens J, Klar L, Hansel C, Slama A, Hager T, Jendrossek V, Aigner C, Klein D. Steens J, et al. Stem Cells Transl Med. 2021 Jan;10(1):128-143. doi: 10.1002/sctm.20-0191. Epub 2020 Aug 24. Stem Cells Transl Med. 2021. PMID: 32830458 Free PMC article. - Reprogramming of tumor-associated macrophages by targeting β-catenin/FOSL2/ARID5A signaling: A potential treatment of lung cancer.
Sarode P, Zheng X, Giotopoulou GA, Weigert A, Kuenne C, Günther S, Friedrich A, Gattenlöhner S, Stiewe T, Brüne B, Grimminger F, Stathopoulos GT, Pullamsetti SS, Seeger W, Savai R. Sarode P, et al. Sci Adv. 2020 Jun 5;6(23):eaaz6105. doi: 10.1126/sciadv.aaz6105. eCollection 2020 Jun. Sci Adv. 2020. PMID: 32548260 Free PMC article. - Cancer stem cell marker CD90 inhibits ovarian cancer formation via β3 integrin.
Chen WC, Hsu HP, Li CY, Yang YJ, Hung YH, Cho CY, Wang CY, Weng TY, Lai MD. Chen WC, et al. Int J Oncol. 2016 Nov;49(5):1881-1889. doi: 10.3892/ijo.2016.3691. Epub 2016 Sep 15. Int J Oncol. 2016. PMID: 27633757 Free PMC article. - Analysis of Intra-Tumoral Macrophages and T Cells in Non-Small Cell Lung Cancer (NSCLC) Indicates a Role for Immune Checkpoint and CD200-CD200R Interactions.
Tøndell A, Subbannayya Y, Wahl SGF, Flatberg A, Sørhaug S, Børset M, Haug M. Tøndell A, et al. Cancers (Basel). 2021 Apr 9;13(8):1788. doi: 10.3390/cancers13081788. Cancers (Basel). 2021. PMID: 33918618 Free PMC article.
References
- Siegel R, Naishadham D, Jemal A (2012) Cancer statistics, 2012. CA Cancer J Clin 62: 10–29. - PubMed
- Ramalingam SS, Owonikoko TK, Khuri FR (2011) Lung cancer: New biological insights and recent therapeutic advances. CA Cancer J Clin 61: 91–112. - PubMed
- Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, et al. (2002) Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med 8: 816–824. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
The authors work was supported by the OTKA PD 83154 grant, by the Predict project (grant no. 259303 of the EU Health.2010.2.4.1.-8 call) and by the KTIA U_BONUS_12-1-2013-0003 grant. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous