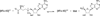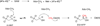Initial characterization of Fom3 from Streptomyces wedmorensis: The methyltransferase in fosfomycin biosynthesis - PubMed (original) (raw)
Initial characterization of Fom3 from Streptomyces wedmorensis: The methyltransferase in fosfomycin biosynthesis
Kylie D Allen et al. Arch Biochem Biophys. 2014.
Abstract
Fosfomycin is a broad-spectrum antibiotic that is useful against multi-drug resistant bacteria. Although its biosynthesis was first studied over 40 years ago, characterization of the penultimate methyl transfer reaction has eluded investigators. The enzyme believed to catalyze this reaction, Fom3, has been identified as a radical S-adenosyl-L-methionine (SAM) superfamily member. Radical SAM enzymes use SAM and a four-iron, four-sulfur ([4Fe-4S]) cluster to catalyze complex chemical transformations. Fom3 also belongs to a family of radical SAM enzymes that contain a putative cobalamin-binding motif, suggesting that it uses cobalamin for methylation. Here we describe the first biochemical characterization of Fom3 from Streptomyces wedmorensis. Since recombinant Fom3 is insoluble, we developed a successful refolding and iron-sulfur cluster reconstitution procedure. Spectroscopic analyses demonstrate that Fom3 binds a [4Fe-4S] cluster which undergoes a transition between a +2 "resting" state and a +1 active state characteristic of radical SAM enzymes. Site-directed mutagenesis of the cysteine residues in the radical SAM CxxxCxxC motif indicates that each residue is essential for functional cluster formation. We also provide preliminary evidence that Fom3 adds a methyl group to 2-hydroxyethylphosphonate (2-HEP) to form 2-hydroxypropylphosphonate (2-HPP) in an apparently SAM-, sodium dithionite-, and methylcobalamin-dependent manner.
Keywords: Cobalamin; Fosfomycin; Iron–sulfur; Methylation; Radical S-adenosyl-l-methionine (SAM); Streptomyces.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
Figure 1
EPR spectra of His-Fom3. A: Bottom, His-Fom3 as purified,Middle, His- Fom3 + DTT, Top, His-Fom3 + DTT + 40 mM dithionite. B: Bottom, His-Fom3 + DTT + 20 mM dithionite,Middle, His-Fom3 + DTT + 20 mM dithionite + SAM,Top, His-Fom3 + DTT + 20 mM dithionite + SAM + 2-HEP + CH3-Cbl(III).
Figure 2
EPR spectra of His-Fom3 variants. Panels A: C282A,B: C286A, C: C289A, **D:**C282A/C286A/C289A. Bottom spectrum (A-D.): protein only. Top spectrum (A-D).: protein + dithionite.
Figure 3
31P NMR spectra, pD ~8. A: Control spectrum of 2-HEP and 2-HPP; B and inset: His-Fom3 in vitro methylation activity; the signal at 0 ppm corresponds to the phosphate of CH3-Cbl(III).
Scheme 1
Fosfomycin biosynthetic pathway in S. wedmorensis.
Scheme 2
SAM reductive cleavage catalyzed by radical SAM enzymes.
Scheme 3
Hypothesized Fom3 reaction mechanism.
Similar articles
- Stereospecific Radical-Mediated B12-Dependent Methyl Transfer by the Fosfomycin Biosynthesis Enzyme Fom3.
McLaughlin MI, van der Donk WA. McLaughlin MI, et al. Biochemistry. 2018 Aug 21;57(33):4967-4971. doi: 10.1021/acs.biochem.8b00616. Epub 2018 Jul 10. Biochemistry. 2018. PMID: 29969250 Free PMC article. - Characterization of the cobalamin-dependent radical S-adenosyl-l-methionine enzyme C-methyltransferase Fom3 in fosfomycin biosynthesis.
Sato S, Kudo F, Eguchi T. Sato S, et al. Methods Enzymol. 2022;669:45-70. doi: 10.1016/bs.mie.2021.11.025. Epub 2021 Dec 31. Methods Enzymol. 2022. PMID: 35644180 - Stereochemical and Mechanistic Investigation of the Reaction Catalyzed by Fom3 from Streptomyces fradiae, a Cobalamin-Dependent Radical S-Adenosylmethionine Methylase.
Wang B, Blaszczyk AJ, Knox HL, Zhou S, Blaesi EJ, Krebs C, Wang RX, Booker SJ. Wang B, et al. Biochemistry. 2018 Aug 21;57(33):4972-4984. doi: 10.1021/acs.biochem.8b00693. Epub 2018 Aug 9. Biochemistry. 2018. PMID: 30036047 Free PMC article. - Radical-mediated enzymatic methylation: a tale of two SAMS.
Zhang Q, van der Donk WA, Liu W. Zhang Q, et al. Acc Chem Res. 2012 Apr 17;45(4):555-64. doi: 10.1021/ar200202c. Epub 2011 Nov 18. Acc Chem Res. 2012. PMID: 22097883 Free PMC article. Review. - S-adenosylmethionine as an oxidant: the radical SAM superfamily.
Wang SC, Frey PA. Wang SC, et al. Trends Biochem Sci. 2007 Mar;32(3):101-10. doi: 10.1016/j.tibs.2007.01.002. Epub 2007 Feb 8. Trends Biochem Sci. 2007. PMID: 17291766 Review.
Cited by
- Consecutive radical S-adenosylmethionine methylations form the ethyl side chain in thienamycin biosynthesis.
Marous DR, Lloyd EP, Buller AR, Moshos KA, Grove TL, Blaszczyk AJ, Booker SJ, Townsend CA. Marous DR, et al. Proc Natl Acad Sci U S A. 2015 Aug 18;112(33):10354-8. doi: 10.1073/pnas.1508615112. Epub 2015 Aug 3. Proc Natl Acad Sci U S A. 2015. PMID: 26240322 Free PMC article. - Identification of a unique radical S-adenosylmethionine methylase likely involved in methanopterin biosynthesis in Methanocaldococcus jannaschii.
Allen KD, Xu H, White RH. Allen KD, et al. J Bacteriol. 2014 Sep;196(18):3315-23. doi: 10.1128/JB.01903-14. Epub 2014 Jul 7. J Bacteriol. 2014. PMID: 25002541 Free PMC article. - Spectroscopic characterization and mechanistic investigation of P-methyl transfer by a radical SAM enzyme from the marine bacterium Shewanella denitrificans OS217.
Allen KD, Wang SC. Allen KD, et al. Biochim Biophys Acta. 2014 Dec;1844(12):2135-44. doi: 10.1016/j.bbapap.2014.09.009. Epub 2014 Sep 16. Biochim Biophys Acta. 2014. PMID: 25224746 Free PMC article. - The B12-Radical SAM Enzyme PoyC Catalyzes Valine Cβ-Methylation during Polytheonamide Biosynthesis.
Parent A, Guillot A, Benjdia A, Chartier G, Leprince J, Berteau O. Parent A, et al. J Am Chem Soc. 2016 Dec 7;138(48):15515-15518. doi: 10.1021/jacs.6b06697. Epub 2016 Nov 29. J Am Chem Soc. 2016. PMID: 27934015 Free PMC article. - Characterization of Two Late-Stage Enzymes Involved in Fosfomycin Biosynthesis in Pseudomonads.
Olivares P, Ulrich EC, Chekan JR, van der Donk WA, Nair SK. Olivares P, et al. ACS Chem Biol. 2017 Feb 17;12(2):456-463. doi: 10.1021/acschembio.6b00939. Epub 2016 Dec 27. ACS Chem Biol. 2017. PMID: 27977135 Free PMC article.
References
- Hidaka T, Goda M, Kuzuyama T, Takei N, Hidaka M, Seto H. Mol Gen Genet. 1995;249:274–280. - PubMed
- Shoji J, Kato T, Hinoo H, Hattori T, Hirooka K, Matsumoto K, Tanimoto T, Kondo E. J Antibiot (Tokyo) 1986;39:1011–1012. - PubMed
- Schito GC. Int J Antimicrob Agents. 2003;22(Suppl 2):79–83. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources