NOTCH1-RBPJ complexes drive target gene expression through dynamic interactions with superenhancers - PubMed (original) (raw)
NOTCH1-RBPJ complexes drive target gene expression through dynamic interactions with superenhancers
Hongfang Wang et al. Proc Natl Acad Sci U S A. 2014.
Abstract
The main oncogenic driver in T-lymphoblastic leukemia is NOTCH1, which activates genes by forming chromatin-associated Notch transcription complexes. Gamma-secretase-inhibitor treatment prevents NOTCH1 nuclear localization, but most genes with NOTCH1-binding sites are insensitive to gamma-secretase inhibitors. Here, we demonstrate that fewer than 10% of NOTCH1-binding sites show dynamic changes in NOTCH1 occupancy when T-lymphoblastic leukemia cells are toggled between the Notch-on and -off states with gamma-secretase inhibiters. Dynamic NOTCH1 sites are functional, being highly associated with Notch target genes, are located mainly in distal enhancers, and frequently overlap with RUNX1 binding. In line with the latter association, we show that expression of IL7R, a gene with key roles in normal T-cell development and in T-lymphoblastic leukemia, is coordinately regulated by Runx factors and dynamic NOTCH1 binding to distal enhancers. Like IL7R, most Notch target genes and associated dynamic NOTCH1-binding sites cooccupy chromatin domains defined by constitutive binding of CCCTC binding factor, which appears to restrict the regulatory potential of dynamic NOTCH1 sites. More remarkably, the majority of dynamic NOTCH1 sites lie in superenhancers, distal elements with exceptionally broad and high levels of H3K27ac. Changes in Notch occupancy produces dynamic alterations in H3K27ac levels across the entire breadth of superenhancers and in the promoters of Notch target genes. These findings link regulation of superenhancer function to NOTCH1, a master regulatory factor and potent oncoprotein in the context of immature T cells, and delineate a generally applicable roadmap for identifying functional Notch sites in cellular genomes.
Keywords: Notch signaling; gene regulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
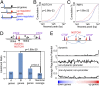
NOTCH1 activation reveals dynamic NOTCH1–RBPJ-binding sites. (A) Scatterplot of NOTCH1 ChIP-seq read counts. Each dot represents a NOTCH1 peak identified in CUTLL1 T-LL cells following GSI washout. ChIP-seq reads within 600 bp of a peak summit were counted in the Notch-on and Notch-off states. Red dots indicate dynamic sites. (B) Classes of NOTCH1-binding sites defined by genomic location and dynamism. Each dot is a NOTCH1 peak plotted according to its distance to the nearest gene’s transcriptional start site (TSS) and its signal-fold change from the Notch-off to the Notch-on states. The red vertical line separates proximal promoters and distal enhancers, and the red horizontal line corresponds to a signal-fold-change threshold with an FDR < 0.05. Inset numbers correspond to peaks found in each quadrant. (C) Heat map of dynamic NOTCH1 sites, ranked by ChIP-Seq signal intensity, and associated transcription factor and histone mark signals across a 1-kb window centered on NOTCH1-binding summits.
Fig. 2.
Identification and functional characterization of the IL7R 3′ enhancers. (A) Chromatin landscapes around the IL7R locus in human T-LL cells shows the presence of a pair of 3′ enhancers, E5 and E3, each containing a dynamic NOTCH1–RBPJ site and a RUNX1 site. (B) Diagram showing IL7R enhancer reporter constructs. Regions spanning E5, E3, or both (E53) NOTCH1–RBPJ binding sites were cloned into the pGL3-TATA box plasmid. (C) IL7R enhancer elements are active in CUTLL1 T-LL cells. Here and elsewhere, luciferase assays were carried out in triplicate and normalized to the luciferase activity generated by the empty pGL3-TATA box plasmid. (D) _IL7R_-enhancer activity in CUTLL1 T-LL cells depends on Notch and Runx factors. Notch and Runx factor activity was inhibited by transfection of plasmids encoding DN-MAML and Runt, respectively. (E) Effects of RBPJ and Runx motif mutations on IL7R E53-enhancer reporter gene activity in CUTLL1 cells. The cartoon shows the IL7R E53-enhancer construct and associated RBPJ and Runx motifs. The effects of mutations involving these sites, alone and in combination, are shown in the IL7R E53-enhancer reporter gene assay below. Error bars in C–E represent 1 sd from the mean of data points obtained in triplicate.
Fig. 3.
Target gene activation through dynamic NOTCH1-binding sites. (A–C) Dynamic NOTCH1–RBPJ-binding sites are preferentially located near genes that are up-regulated by Notch. The distance from the nearest binding site to the TSS of each gene was recorded (A) and cumulative distributions of 340 up-regulated genes (red), 187 down-regulated genes (blue), and the genomic background (black) were plotted for dynamic NOTCH1 (B) and dynamic RBPJ sites (C). P values were calculated by the Kolmogorov–Smirnov test. (D) Constitutive CTCF-binding sites define domains that restrict NOTCH1 regulation of nearby genes. Genes near dynamic NOTCH1 sites were classified into three categories: A, within 100 kb but in a different CTCF domain; B, within 100 kb and in the same CTCF domain; C, more than 100 kb away and in the same CTCF domain. The ratio between the likelihood of finding an activated gene in each category and the likelihood of finding an activated gene randomly are shown in the bar plot; numbers in each bar correspond to the activated genes in each category. P values were calculated by Fisher exact test. (E) Relationships between regulatory potentials and Notch-dependent changes in gene expression. The schematic shows how regulatory potential is calculated (see Materials and Methods for details). The lower panels show relationships between regulatory potentials of NOTCH1-binding sites and differential expression of genes (see
Supporting Information
for details).
Fig. 4.
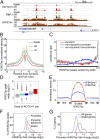
Dynamic NOTCH1 sites are preferentially located within H3K27ac-marked superenhancers. (A) Chromatin landscapes near the NOTCH1 target gene NRARP. The flanking EXD3 gene is not expressed in T-LL. (B) Composite profiles of H3K27ac flanking dynamic NOTCH1 sites under steady-state (basal), Notch-off, and Notch-on conditions. (C) Enrichment of dynamic NOTCH1 sites in the broadest H3K27ac peaks. Genomic H3K27ac peaks (n = 35,244) were ranked by peak width and grouped into 176 bins, each containing 200 peaks. H3K27ac peaks associated with dynamic or nondynamic NOTCH1 sites were counted in each bin, and the ratio of (i) the likelihood of NOTCH1 binding to H3K27ac peaks and (ii) the average likelihood NOTCH1 binding to all genomic H3K27ac peaks is plotted for each class of NOTCH1 sites. (D) Distribution of H3K27ac peak width for all H3K27ac peaks (gray), all promoter H3K27ac peaks (cyan), H3K27ac peaks associated with nondynamic promoter NOTCH1 (dark blue), nondynamic nonpromoter NOTCH1 (light blue), and dynamic NOTCH1 (red) is shown. (E) Composite profiles of H3K27ac and H3K4me1 on dynamic NOTCH1 associated H3K27ac peaks (see
Supporting Information
for details). (F) Fold-change distribution of H3K27ac peaks associated with dynamic NOTCH1 sites versus all H3K27ac peaks. Fold change is the ratio of H3K27ac level in the Notch-on and Notch-off states. (G) Fold-change distribution of promoter H3K27ac levels on NOTCH1 target genes and all genes. H3K27ac level is measured as the normalized read count within 2 kb of transcriptional start sites.
Similar articles
- Long-range enhancer activity determines Myc sensitivity to Notch inhibitors in T cell leukemia.
Yashiro-Ohtani Y, Wang H, Zang C, Arnett KL, Bailis W, Ho Y, Knoechel B, Lanauze C, Louis L, Forsyth KS, Chen S, Chung Y, Schug J, Blobel GA, Liebhaber SA, Bernstein BE, Blacklow SC, Liu XS, Aster JC, Pear WS. Yashiro-Ohtani Y, et al. Proc Natl Acad Sci U S A. 2014 Nov 18;111(46):E4946-53. doi: 10.1073/pnas.1407079111. Epub 2014 Nov 4. Proc Natl Acad Sci U S A. 2014. PMID: 25369933 Free PMC article. - Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells.
Wang H, Zou J, Zhao B, Johannsen E, Ashworth T, Wong H, Pear WS, Schug J, Blacklow SC, Arnett KL, Bernstein BE, Kieff E, Aster JC. Wang H, et al. Proc Natl Acad Sci U S A. 2011 Sep 6;108(36):14908-13. doi: 10.1073/pnas.1109023108. Epub 2011 Jul 7. Proc Natl Acad Sci U S A. 2011. PMID: 21737748 Free PMC article. - The common oncogenomic program of NOTCH1 and NOTCH3 signaling in T-cell acute lymphoblastic leukemia.
Choi SH, Severson E, Pear WS, Liu XS, Aster JC, Blacklow SC. Choi SH, et al. PLoS One. 2017 Oct 12;12(10):e0185762. doi: 10.1371/journal.pone.0185762. eCollection 2017. PLoS One. 2017. PMID: 29023469 Free PMC article. - New insights into Notch1 regulation of the PI3K-AKT-mTOR1 signaling axis: targeted therapy of γ-secretase inhibitor resistant T-cell acute lymphoblastic leukemia.
Hales EC, Taub JW, Matherly LH. Hales EC, et al. Cell Signal. 2014 Jan;26(1):149-61. doi: 10.1016/j.cellsig.2013.09.021. Epub 2013 Oct 16. Cell Signal. 2014. PMID: 24140475 Review. - Transcription Factor RBPJ as a Molecular Switch in Regulating the Notch Response.
Giaimo BD, Gagliani EK, Kovall RA, Borggrefe T. Giaimo BD, et al. Adv Exp Med Biol. 2021;1287:9-30. doi: 10.1007/978-3-030-55031-8_2. Adv Exp Med Biol. 2021. PMID: 33034023 Review.
Cited by
- Characterization of the genome-wide TLX1 binding profile in T-cell acute lymphoblastic leukemia.
Durinck K, Van Loocke W, Van der Meulen J, Van de Walle I, Ongenaert M, Rondou P, Wallaert A, de Bock CE, Van Roy N, Poppe B, Cools J, Soulier J, Taghon T, Speleman F, Van Vlierberghe P. Durinck K, et al. Leukemia. 2015 Dec;29(12):2317-27. doi: 10.1038/leu.2015.162. Epub 2015 Jun 25. Leukemia. 2015. PMID: 26108691 - Novel function of MDA-9/Syntenin (SDCBP) as a regulator of survival and stemness in glioma stem cells.
Talukdar S, Das SK, Pradhan AK, Emdad L, Shen XN, Windle JJ, Sarkar D, Fisher PB. Talukdar S, et al. Oncotarget. 2016 Aug 23;7(34):54102-54119. doi: 10.18632/oncotarget.10851. Oncotarget. 2016. PMID: 27472461 Free PMC article. - Asynchronous combinatorial action of four regulatory factors activates Bcl11b for T cell commitment.
Kueh HY, Yui MA, Ng KK, Pease SS, Zhang JA, Damle SS, Freedman G, Siu S, Bernstein ID, Elowitz MB, Rothenberg EV. Kueh HY, et al. Nat Immunol. 2016 Aug;17(8):956-65. doi: 10.1038/ni.3514. Epub 2016 Jul 4. Nat Immunol. 2016. PMID: 27376470 Free PMC article. - SpDamID: Marking DNA Bound by Protein Complexes Identifies Notch-Dimer Responsive Enhancers.
Hass MR, Liow HH, Chen X, Sharma A, Inoue YU, Inoue T, Reeb A, Martens A, Fulbright M, Raju S, Stevens M, Boyle S, Park JS, Weirauch MT, Brent MR, Kopan R. Hass MR, et al. Mol Cell. 2015 Aug 20;59(4):685-97. doi: 10.1016/j.molcel.2015.07.008. Epub 2015 Aug 6. Mol Cell. 2015. PMID: 26257285 Free PMC article. - Super-enhancers: Asset management in immune cell genomes.
Witte S, O'Shea JJ, Vahedi G. Witte S, et al. Trends Immunol. 2015 Sep;36(9):519-26. doi: 10.1016/j.it.2015.07.005. Epub 2015 Aug 12. Trends Immunol. 2015. PMID: 26277449 Free PMC article. Review.
References
- Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–271. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 HL007627/HL/NHLBI NIH HHS/United States
- R01 GM099409/GM/NIGMS NIH HHS/United States
- P01 CA119070/CA/NCI NIH HHS/United States
- T32HL007627/HL/NHLBI NIH HHS/United States
- R01 AI047833/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases