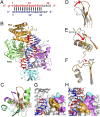Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage - PubMed (original) (raw)
Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage
Gang Sheng et al. Proc Natl Acad Sci U S A. 2014.
Abstract
We report on crystal structures of ternary Thermus thermophilus Argonaute (TtAgo) complexes with 5'-phosphorylated guide DNA and a series of DNA targets. These ternary complex structures of cleavage-incompatible, cleavage-compatible, and postcleavage states solved at improved resolution up to 2.2 Å have provided molecular insights into the orchestrated positioning of catalytic residues, a pair of Mg(2+) cations, and the putative water nucleophile positioned for in-line attack on the cleavable phosphate for TtAgo-mediated target cleavage by a RNase H-type mechanism. In addition, these ternary complex structures have provided insights into protein and DNA conformational changes that facilitate transition between cleavage-incompatible and cleavage-compatible states, including the role of a Glu finger in generating a cleavage-competent catalytic Asp-Glu-Asp-Asp tetrad. Following cleavage, the seed segment forms a stable duplex with the complementary segment of the target strand.
Keywords: DNA guide-DNA target; bacterial Argonaute; catalytic mechanism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
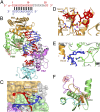
Fig. 1.
Crystal structure and interactions in the _Tt_Ago ternary complex with 5′-phosphorylated 21-mer guide DNA and 12-mer target DNA complementary to segment 2–12 of the guide strand in Mg2+-containing solution. (A) The sequence and pairing of guide (red) and target (blue) strands in the ternary complex. Disordered segments are shown in gray. (B) A 2.9-Å crystal structure of the complex. The various domains and linkers of _Tt_Ago are color-coded, as are the guide and target strands. The catalytic residues in a stick representation are highlighted in a red-dotted background. (C) A view of the guide-target segment highlighting splaying out of 1 and 1′ bases in their respective Mid and PIWI pockets in the complex. The 2–2′ base pair stacks over side chains of Arg446 and His445. (D) Positioning of the 5′-phosphate and sequence-specific recognition of splayed-out base T1 of the guide strand in the Mid pocket. (E) Positioning and sequence-specific recognition of the splayed-out base G1′ of the target strand within a pocket in the PIWI domain. (F) Positioning of Glu512 outside and far away from the catalytic pocket composed of Asp478, Asp546, and Asp660 residues in the ternary complex.
Fig. 2.
Crystal structure and interactions in the _Tt_Ago ternary complex with 5′-phosphorylated 21-mer guide DNA and 19-mer target DNA complementary to segments 2–19 of the guide strand in Mg2+-containing solution. (A) The sequence and pairing of guide (red) and target (blue) strands in the ternary complex. (B) A 2.2-Å crystal structure of the complex. (C) Insertion of Glu512 into the catalytic pocket composed of Asp478, Asp546, and Asp660 residues in the ternary complex. (D_–_F) Conformational changes in loop L1 (D), in loop L2 that contains Glu512 (E), and in loop L3 that contains Asp546 (F) on proceeding from the cleavage-incompatible ternary complex with 12-mer target DNA to the cleavage-compatible ternary complex with 19-mer target DNA. (G and H) Relative positioning of loops L1 (in gold), L2 (in magenta), and L3 (in cyan) in a surface representation on proceeding from the cleavage-incompatible ternary complex with 12-mer target DNA (G) to the cleavage-compatible ternary complex with 19-mer target DNA (H).
Fig. 3.
Structural insights from studies of _Tt_Ago ternary complexes with 5′-phosphorylated 21-mer guide DNA and added 19-mer target DNA in the presence of Mg2+ and Mn2+-containing solution. (A) Intermolecular contacts in the 2.2-Å ternary complex with cleavage-compatible 19-mer target DNA in Mg2+-containing solution. The interactions highlighted by a yellow background are additional contacts observed beyond those observed in the ternary complex with cleavage-incompatible 15-mer target DNA (
SI Appendix, Fig. S5_B_
). (B) Stereoview of the catalytic pocket in the ternary complex with an intact 10′–11′ step on the target strand in Mg2+-containing solution. The pair of Mg2+ cations are labeled “A” and “B” and are shown as magenta balls. Water molecules are shown as pink balls. The four catalytic Asp478, Asp546, Asp660, and Glu512 are shown in stick representation. (C) Stereoview of the catalytic pocket in the ternary complex with a cleaved 10′–11′ step on the target strand in Mg2+-containing solution. (D) A 2.4-Å crystal structure and interactions in the _Tt_Ago ternary complex with 5′-phosphorylated 21-mer guide DNA and cleaved 19-mer target DNA complementary to segment 2–19 of the guide strand for crystals grown in Mn2+ containing solution. The target DNA is cleaved at the 10′–11′ step on the DNA target strand. The Inset expands the catalytic pocket segment showing the cleavage of the backbone.
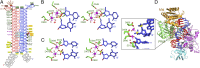
Fig. 4.
Crystal structure and interactions in the _Tt_Ago ternary complex with 5′-phosphorylated 21-mer guide DNA and 15-mer target DNA complementary to segments 2–15 of the guide strand in Mg2+-containing solution. (A) The sequence and pairing of guide (red) and target (blue) strands in the ternary complex. (B) A 2.25-Å crystal structure of the complex. (C) Positioning of Glu512 outside and far away from the catalytic pocket in the ternary complex with 15-mer target DNA. (D) Insertion of Glu512 into the catalytic pocket in the ternary complex with 15-mer target RNA reported previously (Protein Data Bank ID code 3HJF). (E) Superposition of the guide DNA-target DNA (in silver) and guide DNA-target RNA (in magneta) in the _Tt_Ago ternary complexes containing 15-mer target DNAs and RNAs. Note that we observed one more base pair (15–15′) in the ternary complex with target RNA. (F) Superposition of the catalytic pockets and loops L1, L2, and L3 in the ternary complexes with target DNA (in silver) and target RNA (in magenta).
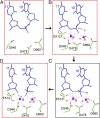
Fig. 5.
A proposed mechanism for Ago-mediated Mg2+ cation-dependent cleavage of target strands. Crystal structure snapshots (A, B, and D) and a proposed model of transition state (C) in the reaction pathway leading to cleavage of the target DNA strand at the 10′–11′ step in the ternary complex of _Tt_Ago with complementary guide and target DNA strands. (A) Structure of the catalytic pocket in the cleavage-incompatible ternary complex with Glu512 positioned outside and far from the catalytic pocket as observed in ternary complexes with 12- and 15-mer target DNA strands. (B) Structure of the catalytic pocket in the cleavage-compatible ternary complex with Gln512 inserted into the catalytic pocket as observed in the ternary complex with 16- and 19-mer target DNA strands. (C) Proposed model of the transition state of the cleavage reaction in the ternary complex. (D) Structure of the catalytic pocket of the ternary complex following cleavage of the 10′–11′ backbone in the ternary complex with cleaved 19-mer target DNA strand.
Similar articles
- Two symmetric arginine residues play distinct roles in Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Lei J, Sheng G, Cheung PP, Wang S, Li Y, Gao X, Zhang Y, Wang Y, Huang X. Lei J, et al. Proc Natl Acad Sci U S A. 2019 Jan 15;116(3):845-853. doi: 10.1073/pnas.1817041116. Epub 2018 Dec 27. Proc Natl Acad Sci U S A. 2019. PMID: 30591565 Free PMC article. - Structure/cleavage-based insights into helical perturbations at bulge sites within T. thermophilus Argonaute silencing complexes.
Sheng G, Gogakos T, Wang J, Zhao H, Serganov A, Juranek S, Tuschl T, Patel DJ, Wang Y. Sheng G, et al. Nucleic Acids Res. 2017 Sep 6;45(15):9149-9163. doi: 10.1093/nar/gkx547. Nucleic Acids Res. 2017. PMID: 28911094 Free PMC article. - Gene Silencing Mechanisms Revealed by Dynamics of Guide, Target, and Duplex Binding to Argonaute.
Tai HC, Lim C. Tai HC, et al. J Chem Theory Comput. 2020 Jan 14;16(1):688-699. doi: 10.1021/acs.jctc.9b00546. Epub 2019 Dec 10. J Chem Theory Comput. 2020. PMID: 31751512 - Understanding the core of RNA interference: The dynamic aspects of Argonaute-mediated processes.
Zhu L, Jiang H, Sheong FK, Cui X, Wang Y, Gao X, Huang X. Zhu L, et al. Prog Biophys Mol Biol. 2017 Sep;128:39-46. doi: 10.1016/j.pbiomolbio.2016.09.008. Epub 2016 Sep 30. Prog Biophys Mol Biol. 2017. PMID: 27697475 Review. - The evolutionary journey of Argonaute proteins.
Swarts DC, Makarova K, Wang Y, Nakanishi K, Ketting RF, Koonin EV, Patel DJ, van der Oost J. Swarts DC, et al. Nat Struct Mol Biol. 2014 Sep;21(9):743-53. doi: 10.1038/nsmb.2879. Nat Struct Mol Biol. 2014. PMID: 25192263 Free PMC article. Review.
Cited by
- The Slicer Activity of ARGONAUTE1 Is Required Specifically for the Phasing, Not Production, of Trans-Acting Short Interfering RNAs in Arabidopsis.
Arribas-Hernández L, Marchais A, Poulsen C, Haase B, Hauptmann J, Benes V, Meister G, Brodersen P. Arribas-Hernández L, et al. Plant Cell. 2016 Jul;28(7):1563-80. doi: 10.1105/tpc.16.00121. Epub 2016 Jun 27. Plant Cell. 2016. PMID: 27354557 Free PMC article. - A conditional protein diffusion model generates artificial programmable endonuclease sequences with enhanced activity.
Zhou B, Zheng L, Wu B, Yi K, Zhong B, Tan Y, Liu Q, Liò P, Hong L. Zhou B, et al. Cell Discov. 2024 Sep 10;10(1):95. doi: 10.1038/s41421-024-00728-2. Cell Discov. 2024. PMID: 39251570 Free PMC article. - Expansion and Divergence of Argonaute Genes in the Oomycete Genus Phytophthora.
Bollmann SR, Press CM, Tyler BM, Grünwald NJ. Bollmann SR, et al. Front Microbiol. 2018 Nov 30;9:2841. doi: 10.3389/fmicb.2018.02841. eCollection 2018. Front Microbiol. 2018. PMID: 30555430 Free PMC article. - Two symmetric arginine residues play distinct roles in Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Lei J, Sheng G, Cheung PP, Wang S, Li Y, Gao X, Zhang Y, Wang Y, Huang X. Lei J, et al. Proc Natl Acad Sci U S A. 2019 Jan 15;116(3):845-853. doi: 10.1073/pnas.1817041116. Epub 2018 Dec 27. Proc Natl Acad Sci U S A. 2019. PMID: 30591565 Free PMC article. - Applications of Programmable Endonucleases in Sequence- and Ligation-Independent Seamless DNA Assembly.
Xiong X, Lu Z, Ma L, Zhai C. Xiong X, et al. Biomolecules. 2023 Jun 21;13(7):1022. doi: 10.3390/biom13071022. Biomolecules. 2023. PMID: 37509059 Free PMC article. Review.
References
- Peters L, Meister G. Argonaute proteins: Mediators of RNA silencing. Mol Cell. 2007;26(5):611–623. - PubMed
- Hutvagner G, Simard MJ. Argonaute proteins: Key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9(1):22–32. - PubMed
- Kawamata T, Tomari Y. Making RISC. Trends Biochem Sci. 2010;35(7):368–376. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources