Spatial organization of RNA polymerase II inside a mammalian cell nucleus revealed by reflected light-sheet superresolution microscopy - PubMed (original) (raw)
Spatial organization of RNA polymerase II inside a mammalian cell nucleus revealed by reflected light-sheet superresolution microscopy
Ziqing W Zhao et al. Proc Natl Acad Sci U S A. 2014.
Abstract
Superresolution microscopy based on single-molecule centroid determination has been widely applied to cellular imaging in recent years. However, quantitative imaging of the mammalian nucleus has been challenging due to the lack of 3D optical sectioning methods for normal-sized cells, as well as the inability to accurately count the absolute copy numbers of biomolecules in highly dense structures. Here we report a reflected light-sheet superresolution microscopy method capable of imaging inside the mammalian nucleus with superior signal-to-background ratio as well as molecular counting with single-copy accuracy. Using reflected light-sheet superresolution microscopy, we probed the spatial organization of transcription by RNA polymerase II (RNAP II) molecules and quantified their global extent of clustering inside the mammalian nucleus. Spatiotemporal clustering analysis that leverages on the blinking photophysics of specific organic dyes showed that the majority (>70%) of the transcription foci originate from single RNAP II molecules, and no significant clustering between RNAP II molecules was detected within the length scale of the reported diameter of "transcription factories." Colocalization measurements of RNAP II molecules equally labeled by two spectrally distinct dyes confirmed the primarily unclustered distribution, arguing against a prevalent existence of transcription factories in the mammalian nucleus as previously proposed. The methods developed in our study pave the way for quantitative mapping and stoichiometric characterization of key biomolecular species deep inside mammalian cells.
Keywords: intracellular molecular counting; mammalian gene transcription; nuclear organization; quantitative fluorescence microscopy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
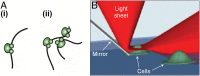
“Transcription factories” hypothesis and RLS-SRM imaging. (A) Models of spatial distribution of RNAP II in the mammalian nucleus, in which transcription is carried out by either (i) individual RNAP II molecules or (ii) multiple molecules clustered into spatially discrete “factories” that pull together genes to be transcribed. (B) The principle of RLS-SRM, which uses a miniature mirror placed next to the cell to reflect a light sheet by 90° to achieve 3D optical sectioning.
Fig. 2.
Molecular counting based on spatiotemporal clustering analysis. (A) Comparison of blinking kinetics between TMR (rhodamine-based, Bottom) and Alexa 647 (cyanine-based, Top) dyes. Representative time traces show that although both dyes remain “on” for similar periods of time, TMR tends to exhibit much longer “off” times, making its blinking trajectories appear clustered in time and allowing temporal clustering to be performed. (B) Flowchart of spatiotemporal clustering analysis based on spatial and temporal NND thresholding of TMR blinking events, which enables accurate counting of RNAP II molecules in transcription foci. (C) Calibration for temporal clustering of TMR localizations using hybridized DNA constructs mimicking transcription foci with one or multiple RNAP II molecules. A plot of the average number of st-clusters per spatial cluster (as determined from spatiotemporal clustering analysis) against the average number of dyes per construct molecule (as determined by counting photobleaching steps) allows us to establish an unambiguous correspondence between the average number of st-clusters observed and the average copy number of RNAP II molecules in the transcription foci. Error bar denotes SD (n = 3).
Fig. 3.
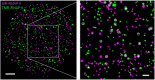
Spatial organization of RNAP II molecules shows no significant clustering. (A) Distribution of SNAP-RPB1 molecules in a thin optical section of the nucleus of a fixed U2OS cell labeled with TMR. (Inset) Zoomed-in area where individual transcription foci are discernible; yellow crosses indicate the centroid position of the st-clusters identified. (Scale bar, 2 μm; Inset, 500 nm.) (B) Distribution of the number of st-clusters in transcription foci indicates that at least 70% of the foci consist of only one RNAP II molecule (n = 4,465). (C) Distribution of spatial NND for transcription foci shows that the majority of the RNAP II molecules do not associate with each other within the reported diameter of transcription factories (40–130 nm). Dotted line indicates the mean.
Fig. 4.
Quantification of RNAP II clustering by two-color colocalization. SNAP-RPB1 molecules are simultaneously labeled with either SiR (cyan) or TMR (green), so that approximately half of the molecules are labeled with each dye. Molecules that colocalize with each other are highlighted with white circles in the Inset. (Scale bar, 2 μm; Inset, 500 nm.)
Similar articles
- Towards a 'Spot On' Understanding of Transcription in the Nucleus.
Patange S, Ball DA, Karpova TS, Larson DR. Patange S, et al. J Mol Biol. 2021 Jul 9;433(14):167016. doi: 10.1016/j.jmb.2021.167016. Epub 2021 May 2. J Mol Biol. 2021. PMID: 33951451 Free PMC article. Review. - Study of RNA Polymerase II Clustering inside Live-Cell Nuclei Using Bayesian Nanoscopy.
Chen X, Wei M, Zheng MM, Zhao J, Hao H, Chang L, Xi P, Sun Y. Chen X, et al. ACS Nano. 2016 Feb 23;10(2):2447-54. doi: 10.1021/acsnano.5b07257. Epub 2016 Feb 11. ACS Nano. 2016. PMID: 26855123 - Dual-color 3D-dSTORM colocalization and quantification of ROXY1 and RNAPII variants throughout the transcription cycle in root meristem nuclei.
Maß L, Holtmannspötter M, Zachgo S. Maß L, et al. Plant J. 2020 Dec;104(5):1423-1436. doi: 10.1111/tpj.14986. Epub 2020 Oct 5. Plant J. 2020. PMID: 32896918 - Imaging transcription elongation dynamics by new technologies unveils the organization of initiation and elongation in transcription factories.
Kimura H, Sato Y. Kimura H, et al. Curr Opin Cell Biol. 2022 Feb;74:71-79. doi: 10.1016/j.ceb.2022.01.002. Epub 2022 Feb 17. Curr Opin Cell Biol. 2022. PMID: 35183895 Review. - How Single-Molecule Localization Microscopy Expanded Our Mechanistic Understanding of RNA Polymerase II Transcription.
Hoboth P, Šebesta O, Hozák P. Hoboth P, et al. Int J Mol Sci. 2021 Jun 22;22(13):6694. doi: 10.3390/ijms22136694. Int J Mol Sci. 2021. PMID: 34206594 Free PMC article. Review.
Cited by
- Transcription-Dependent Formation of Nuclear Granules Containing FUS and RNA Pol II.
Thompson VF, Victor RA, Morera AA, Moinpour M, Liu MN, Kisiel CC, Pickrel K, Springhower CE, Schwartz JC. Thompson VF, et al. Biochemistry. 2018 Dec 26;57(51):7021-7032. doi: 10.1021/acs.biochem.8b01097. Epub 2018 Dec 11. Biochemistry. 2018. PMID: 30488693 Free PMC article. - Visualizing transcription factor dynamics in living cells.
Liu Z, Tjian R. Liu Z, et al. J Cell Biol. 2018 Apr 2;217(4):1181-1191. doi: 10.1083/jcb.201710038. Epub 2018 Jan 29. J Cell Biol. 2018. PMID: 29378780 Free PMC article. Review. - Direct visualization of cardiac transcription factories reveals regulatory principles of nuclear architecture during pathological remodeling.
Karbassi E, Rosa-Garrido M, Chapski DJ, Wu Y, Ren S, Wang Y, Stefani E, Vondriska TM. Karbassi E, et al. J Mol Cell Cardiol. 2019 Mar;128:198-211. doi: 10.1016/j.yjmcc.2019.02.003. Epub 2019 Feb 8. J Mol Cell Cardiol. 2019. PMID: 30742811 Free PMC article. - Super-Resolution Microscopy in Studying the Structure and Function of the Cell Nucleus.
Ryabichko SS, Ibragimov AN, Lebedeva LA, Kozlov EN, Shidlovskii YV. Ryabichko SS, et al. Acta Naturae. 2017 Oct-Dec;9(4):42-51. Acta Naturae. 2017. PMID: 29340216 Free PMC article. - Activation of new replication foci under conditions of replication stress.
Rybak P, Waligórska A, Bujnowicz Ł, Hoang A, Dobrucki JW. Rybak P, et al. Cell Cycle. 2015;14(16):2634-47. doi: 10.1080/15384101.2015.1064566. Cell Cycle. 2015. PMID: 26212617 Free PMC article.
References
- Edelman LB, Fraser P. Transcription factories: Genetic programming in three dimensions. Curr Opin Genet Dev. 2012;22(2):110–114. - PubMed
- Papantonis A, Cook PR. Transcription factories: Genome organization and gene regulation. Chem Rev. 2013;113(11):8683–8705. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM096450/GM/NIGMS NIH HHS/United States
- GM 096450/GM/NIGMS NIH HHS/United States
- EB010244/EB/NIBIB NIH HHS/United States
- T32 GM008313/GM/NIGMS NIH HHS/United States
- R01 EB010244/EB/NIBIB NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources