Proteotoxicity: an underappreciated pathology in cardiac disease - PubMed (original) (raw)
Review
Proteotoxicity: an underappreciated pathology in cardiac disease
Marco Sandri et al. J Mol Cell Cardiol. 2014 Jun.
Abstract
In general, in most organ systems, intracellular protein homeostasis is the sum of many factors, including chromosomal state, protein synthesis, post-translational processing and transport, folding, assembly and disassembly into macromolecular complexes, protein stability and clearance. In the heart, there has been a focus on the gene programs that are activated during pathogenic processes, but the removal of damaged proteins and organelles has been underappreciated as playing an important role in the pathogenesis of heart disease. Proteotoxicity refers to the adverse effects of damaged or misfolded proteins and even organelles on the cell. At the cellular level, the ultimate outcome of uncontrolled or severe proteotoxicity is cell death; hence, the pathogenic impact of proteotoxicity is maximally manifested in organs with no or very poor regenerative capability such as the brain and the heart. Evidence for increased cardiac proteotoxicity is rapidly mounting for a large subset of congenital and acquired human heart disease. Studies carried out in animal models and in cell culture have begun to establish both sufficiency and, in some cases, the necessity of proteotoxicity as a major pathogenic factor in the heart. This dictates rigorous testing for the efficacy of proteotoxic attenuation as a new strategy to treat heart disease. This review article highlights some recent advances in our understanding of how misfolded proteins can injure and are handled in the cell, examining the emerging evidence for targeting proteotoxicity as a new therapeutic strategy for heart disease. This article is part of a Special Issue entitled "Protein Quality Control, the Ubiquitin Proteasome System, and Autophagy."
Keywords: Autophagy; Lysosome; Protein.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures
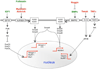
Figure 1
Signalling pathways that control the expression of the ubiquitin ligases in striated muscles. In red are depicted the hormones/cytokines that have a catabolic action while in green are underlined the anabolic ones.
Figure 2
Striated muscle regulation of macroautophagy, microautophagy and chaperone-mediated autophagy (CMA). The different colors underline modules describing the critical steps in the three types of autophagy. The different modules are: autophagosome formation, selective autophagic removal of mitochondria (mitophagy), autophagosome docking, direct glycogen uptake from lysosomes and removal of unfolded/misfolded proteins containing a KFERQ sequence. The dotted lines point to mechanisms that are not yet known. FOXO transcription factors are master regulators of several modules in macroautophagy.
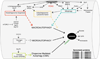
Figure 3
The initial stages of vesicle elongation in macroautophagy. A. Only the initial stages are shown, focusing on the seminal role played by Atg7 in the process, where it intersects with the two possible pathways. The first pathway (top) occurs through the direct interaction of Atg12 with Atg5. This interaction requires the E1-like activity of Atg7. The second pathway involves the addition of phosphotidylethanolamine (PE, shown lower left) to LC3-I, the mammalian homologue of the yeast protein, Atg8. This process is mediated by a protease, Atg4 followed by Atg7’s E1-like enzymatic activity to add PE to LC3-I, coupled with the E2-like activity of Atg3. B. Electron micrograph of a cardiomyocyte derived from the heart of mouse that overexpresses (approximately 10-fold) Atg7. Immediately apparent are elaborated, linear membranes. These structures do not affect cardiac function during the animal’s lifetime [94].
Similar articles
- Priming the Proteasome to Protect against Proteotoxicity.
Wang X, Wang H. Wang X, et al. Trends Mol Med. 2020 Jul;26(7):639-648. doi: 10.1016/j.molmed.2020.02.007. Epub 2020 Mar 26. Trends Mol Med. 2020. PMID: 32589934 Free PMC article. Review. - Proteasomal and lysosomal protein degradation and heart disease.
Wang X, Robbins J. Wang X, et al. J Mol Cell Cardiol. 2014 Jun;71:16-24. doi: 10.1016/j.yjmcc.2013.11.006. Epub 2013 Nov 14. J Mol Cell Cardiol. 2014. PMID: 24239609 Free PMC article. Review. - The interplay between autophagy and the ubiquitin-proteasome system in cardiac proteotoxicity.
Wang C, Wang X. Wang C, et al. Biochim Biophys Acta. 2015 Feb;1852(2):188-94. doi: 10.1016/j.bbadis.2014.07.028. Epub 2014 Aug 1. Biochim Biophys Acta. 2015. PMID: 25092168 Free PMC article. Review. - p62 Stages an interplay between the ubiquitin-proteasome system and autophagy in the heart of defense against proteotoxic stress.
Su H, Wang X. Su H, et al. Trends Cardiovasc Med. 2011 Nov;21(8):224-8. doi: 10.1016/j.tcm.2012.05.015. Trends Cardiovasc Med. 2011. PMID: 22902070 Free PMC article. Review. - The heart of autophagy: deconstructing cardiac proteotoxicity.
Rothermel BA, Hill JA. Rothermel BA, et al. Autophagy. 2008 Oct;4(7):932-5. doi: 10.4161/auto.6756. Epub 2008 Oct 8. Autophagy. 2008. PMID: 18769158 Free PMC article.
Cited by
- Increased clearance of reactive aldehydes and damaged proteins in hypertension-induced compensated cardiac hypertrophy: impact of exercise training.
Campos JC, Fernandes T, Bechara LR, da Paixão NA, Brum PC, de Oliveira EM, Ferreira JC. Campos JC, et al. Oxid Med Cell Longev. 2015;2015:464195. doi: 10.1155/2015/464195. Epub 2015 Apr 14. Oxid Med Cell Longev. 2015. PMID: 25954323 Free PMC article. - Priming the Proteasome to Protect against Proteotoxicity.
Wang X, Wang H. Wang X, et al. Trends Mol Med. 2020 Jul;26(7):639-648. doi: 10.1016/j.molmed.2020.02.007. Epub 2020 Mar 26. Trends Mol Med. 2020. PMID: 32589934 Free PMC article. Review. - Paralogue-Specific Roles of SUMO1 and SUMO2/3 in Protein Quality Control and Associated Diseases.
Wang W, Matunis MJ. Wang W, et al. Cells. 2023 Dec 20;13(1):8. doi: 10.3390/cells13010008. Cells. 2023. PMID: 38201212 Free PMC article. Review. - Hyperactive mTORC1/4EBP1 signaling dysregulates proteostasis and accelerates cardiac aging.
Zarzycka W, Kobak KA, King CJ, Peelor FF 3rd, Miller BF, Chiao YA. Zarzycka W, et al. Geroscience. 2025 Apr;47(2):1823-1836. doi: 10.1007/s11357-024-01368-w. Epub 2024 Oct 9. Geroscience. 2025. PMID: 39379739 Free PMC article. - Breaking down the COP9 Signalsome in the heart: how inactivating a protein ubiquitin ligase increases protein ubiquitylation and protects the heart.
Glembotski CC. Glembotski CC. Circ Res. 2015 Nov 6;117(11):914-6. doi: 10.1161/CIRCRESAHA.115.307644. Circ Res. 2015. PMID: 26541679 Free PMC article. No abstract available.
References
- Willis MS, Patterson C. Proteotoxicity and cardiac dysfunction--Alzheimer's disease of the heart? N Engl J Med. 2013;368:455–464. - PubMed
- Hightower LE. Heat shock, stress proteins, chaperones, and proteotoxicity. Cell. 1991;66:191–197. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL105924/HL/NHLBI NIH HHS/United States
- P01HL077101/HL/NHLBI NIH HHS/United States
- P01 HL069779/HL/NHLBI NIH HHS/United States
- P01 HL059408/HL/NHLBI NIH HHS/United States
- R01HL105924/HL/NHLBI NIH HHS/United States
- P01HL049058/HL/NHLBI NIH HHS/United States
- P50 HL077101/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical