Expression of the autophagy substrate SQSTM1/p62 is restored during prolonged starvation depending on transcriptional upregulation and autophagy-derived amino acids - PubMed (original) (raw)
Expression of the autophagy substrate SQSTM1/p62 is restored during prolonged starvation depending on transcriptional upregulation and autophagy-derived amino acids
Mayurbhai Himatbhai Sahani et al. Autophagy. 2014 Mar.
Abstract
SQSTM1/p62 (sequestosome 1) is a multifunctional signaling molecule, involved in a variety of cellular pathways. SQSTM1 is one of the best-known autophagic substrates, and is therefore widely used as an indicator of autophagic degradation. Here we report that the expression level of SQSTM1 can be restored during prolonged starvation. Upon starvation, SQSTM1 is initially degraded by autophagy. However, SQSTM1 is restored back to basal levels during prolonged starvation in mouse embryonic fibroblasts and HepG2 cells, but not in HeLa and HEK293 cells. Restoration of SQSTM1 depends on its transcriptional upregulation, which is triggered by amino acid starvation. Furthermore, amino acids derived from the autophagy-lysosome pathway are used for de novo synthesis of SQSTM1 under starvation conditions. The restoration of SQSTM1 is independent of reactivation of MTORC1 (mechanistic target of rapamycin complex 1). These results suggest that the expression level of SQSTM1 in starved cells is determined by at least 3 factors: autophagic degradation, transcriptional upregulation, and availability of lysosomal-derived amino acids. The results of this study also indicate that the expression level of SQSTM1 does not always inversely correlate with autophagic activity.
Keywords: SQSTM1/p62; amino acid; transcription.
Figures
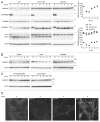
Figure 1. SQSTM1 is restored during prolonged starvation. (A) Wild-type, Rb1cc1 KO and Atg5 KO MEFs were cultured in starvation medium lacking amino acids and serum for 1, 2, 4, 6, and 8 h. Cell lysates were analyzed by immunoblotting using the indicated antibodies. Densitometric quantification of SQSTM1 protein levels using ImageJ software is shown on the graph. Data represent the mean ± SEM of 4 independent experiments (including the data in Figs. 1A, 4A, and 5A) for wild-type cells. A representative result is shown for Rb1cc1 KO and Atg5 KO cells. (B) HepG2, HeLa and HEK293 cells were analyzed as in (A). (C) Wild-type immortalized and primary MEFs were analyzed as in (A). (D) Wild-type MEFs were cultured in starvation medium lacking amino acids and serum for 2, 4, and 8 h. Cells were fixed and stained with anti-SQSTM1/p62 antibody. Scale bar: 5 µm.
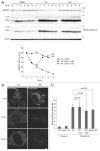
Figure 2. SQSTM1 restoration requires de novo protein synthesis but is independent of MTORC1. Wild-type MEFs were cultured in starvation medium lacking amino acids and serum for 1, 2, 4, 6, and 8 h. At 3 h after starvation, Torin1 (250 nM) or cycloheximide (CHX, 50 μg/ml) was added. Cell lysates were analyzed by immunoblotting using the indicated antibodies. Densitometric quantification of SQSTM1 protein levels using ImageJ software is shown on the graph.
Figure 3. Amino acid starvation-induced upregulation of Sqstm1 transcription is required for SQSTM1 restoration during prolonged starvation. (A) Wild-type MEFs were cultured in DMEM lacking both amino acids and serum, or either amino acids or serum alone, or regular DMEM containing 250 nM Torin1 for 2, 4, and 8 h. Relative Sqstm1 mRNA is estimated by quantitative PCR using Actb as an internal control. Data represent the mean ± SEM of 3 independent experiments. (B) HepG2, HeLa and HEK293 cells were cultured in DMEM lacking both amino acids and serum and analyzed as in (A). (C) Wild-type MEFs were cultured as in (A). The SQSTM1 protein level was analyzed by immunoblotting. ACTB was used as a loading control. Densitometric quantification of SQSTM1 protein levels using ImageJ software is shown on the graph. Data represent the mean ± SEM of 3 independent experiments. (D) Wild-type MEFs stably expressing GFP-SQSTM1 were cultured in DMEM without amino acids and serum. Expression levels of endogenous SQSTM1 and GFP-SQSTM1 were analyzed as in (C). (E) Wild-type MEFs were pretreated with or without 20 µg/ml α-amanitin for 4 h, and then cultured in DMEM without serum and amino acids for 4 or 8 h in the presence or absence of 20 µg/ml α-amanitin. Sqstm1 mRNA was quantified as in (A). (F) Wild-type MEFs were treated as in (E) and the SQSTM1 protein level was analyzed by immunoblotting. ACTB was used as a loading control. Densitometric quantification of SQSTM1 protein levels using ImageJ software is shown on the graph.
Figure 4. Inhibition of lysosomal function abolishes SQSTM1 restoration. (A) Wild-type MEFs were cultured in DMEM lacking both amino acids and serum in the presence of 20 µM chloroquine (CQ) or 0.1 µM bafilomycinA1 (Baf) for 1, 2, 4, 6, and 8 h. Densitometric quantification of SQSTM1 protein levels is shown on the graph. (B) Wild-type MEFs were cultured in regular DMEM medium. These MEFs were pretreated with 1 µM LysoTracker for 30 min and washed with PBS twice. Pretreated MEFs were treated with 20 µM CQ or 0.1 µM Baf for indicated times. Scale bar: 5 µm. (C) Wild-type and Rb1cc1 KO MEFs were cultured in regular DMEM medium or starvation medium (DMEM without amino acids and serum) for 4 h. Wild-type MEFs were also treated with 20 µM CQ or 0.1 µM Baf during the 4 h of starvation. Relative Sqstm1 mRNA was estimated by quantitative PCR. Data represent mean ± SEM of 3 independent experiments.
Figure 5. Amino acid replenishment causes restoration of SQSTM1 in bafilomycinA1-treated cells. (A) Wild-type MEFs were cultured in starvation medium containing 0.1 µM bafilomycinA1 (Baf) for 1, 2, 4, 6, and 8 h. After 3 h of starvation treatment, the medium was replaced with regular DMEM containing amino acids with or without 250 nM Torin1. Densitometric quantification of SQSTM1 protein levels using ImageJ software is shown on the graph. Data represent the mean ± SEM of 3 independent experiments. (B) Wild-type MEFs were cultured in starvation medium containing 20 µM chloroquine (CQ) for 1, 2, 4, 6, and 8 h. After 3 h of starvation treatment, the medium was replaced with regular DMEM containing amino acids with or without 250 nM Torin1.
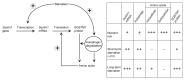
Figure 6. A model of SQSTM1 restoration during prolonged starvation. Under nutrient-rich conditions, SQSTM1 protein is maintained at basal levels. During short-term starvation (~2 h), SQSTM1 is rapidly degraded by autophagy. As starvation-induced transcriptional upregulation of SQSTM1 is only modest, overall SQSTM1 protein levels decrease. Following prolonged starvation, high levels of Sqstm1 mRNA and sufficient amounts of intracellular amino acids derived from autophagy promote de novo synthesis of SQSTM1, which restores SQSTM1 protein expression to basal levels.
Similar articles
- Role of autophagy, SQSTM1, SH3GLB1, and TRIM63 in the turnover of nicotinic acetylcholine receptors.
Khan MM, Strack S, Wild F, Hanashima A, Gasch A, Brohm K, Reischl M, Carnio S, Labeit D, Sandri M, Labeit S, Rudolf R. Khan MM, et al. Autophagy. 2014 Jan;10(1):123-36. doi: 10.4161/auto.26841. Epub 2013 Nov 8. Autophagy. 2014. PMID: 24220501 Free PMC article. - LC3- and p62-based biochemical methods for the analysis of autophagy progression in mammalian cells.
Jiang P, Mizushima N. Jiang P, et al. Methods. 2015 Mar;75:13-8. doi: 10.1016/j.ymeth.2014.11.021. Epub 2014 Dec 5. Methods. 2015. PMID: 25484342 - p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death.
Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. Bjørkøy G, et al. J Cell Biol. 2005 Nov 21;171(4):603-14. doi: 10.1083/jcb.200507002. Epub 2005 Nov 14. J Cell Biol. 2005. PMID: 16286508 Free PMC article. - p62/SQSTM1 functions as a signaling hub and an autophagy adaptor.
Katsuragi Y, Ichimura Y, Komatsu M. Katsuragi Y, et al. FEBS J. 2015 Dec;282(24):4672-8. doi: 10.1111/febs.13540. Epub 2015 Oct 16. FEBS J. 2015. PMID: 26432171 Review. - P62/SQSTM1 at the interface of aging, autophagy, and disease.
Bitto A, Lerner CA, Nacarelli T, Crowe E, Torres C, Sell C. Bitto A, et al. Age (Dordr). 2014 Jun;36(3):9626. doi: 10.1007/s11357-014-9626-3. Epub 2014 Feb 21. Age (Dordr). 2014. PMID: 24557832 Free PMC article. Review.
Cited by
- Small-molecule enhancers of autophagy modulate cellular disease phenotypes suggested by human genetics.
Kuo SY, Castoreno AB, Aldrich LN, Lassen KG, Goel G, Dančík V, Kuballa P, Latorre I, Conway KL, Sarkar S, Maetzel D, Jaenisch R, Clemons PA, Schreiber SL, Shamji AF, Xavier RJ. Kuo SY, et al. Proc Natl Acad Sci U S A. 2015 Aug 4;112(31):E4281-7. doi: 10.1073/pnas.1512289112. Epub 2015 Jul 20. Proc Natl Acad Sci U S A. 2015. PMID: 26195741 Free PMC article. - Loss of VAPB Regulates Autophagy in a Beclin 1-Dependent Manner.
Wu D, Hao Z, Ren H, Wang G. Wu D, et al. Neurosci Bull. 2018 Dec;34(6):1037-1046. doi: 10.1007/s12264-018-0276-9. Epub 2018 Aug 24. Neurosci Bull. 2018. PMID: 30143980 Free PMC article. - Libertellenone T, a Novel Compound Isolated from Endolichenic Fungus, Induces G2/M Phase Arrest, Apoptosis, and Autophagy by Activating the ROS/JNK Pathway in Colorectal Cancer Cells.
Gamage CDB, Kim JH, Yang Y, Taş İ, Park SY, Zhou R, Pulat S, Varlı M, Hur JS, Nam SJ, Kim H. Gamage CDB, et al. Cancers (Basel). 2023 Jan 12;15(2):489. doi: 10.3390/cancers15020489. Cancers (Basel). 2023. PMID: 36672439 Free PMC article. - Rainbow Trout Red Blood Cells Exposed to Viral Hemorrhagic Septicemia Virus Up-Regulate Antigen-Processing Mechanisms and MHC I&II, CD86, and CD83 Antigen-presenting Cell Markers.
Nombela I, Requena-Platek R, Morales-Lange B, Chico V, Puente-Marin S, Ciordia S, Mena MC, Coll J, Perez L, Mercado L, Ortega-Villaizan MDM. Nombela I, et al. Cells. 2019 Apr 27;8(5):386. doi: 10.3390/cells8050386. Cells. 2019. PMID: 31035565 Free PMC article. - Preventing mutant huntingtin proteolysis and intermittent fasting promote autophagy in models of Huntington disease.
Ehrnhoefer DE, Martin DDO, Schmidt ME, Qiu X, Ladha S, Caron NS, Skotte NH, Nguyen YTN, Vaid K, Southwell AL, Engemann S, Franciosi S, Hayden MR. Ehrnhoefer DE, et al. Acta Neuropathol Commun. 2018 Mar 6;6(1):16. doi: 10.1186/s40478-018-0518-0. Acta Neuropathol Commun. 2018. PMID: 29510748 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources