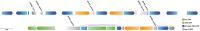Genome degeneration and adaptation in a nascent stage of symbiosis - PubMed (original) (raw)
Rosario Gil, Adam L Clayton, Diane M Dunn, Andrew C von Niederhausern, Cindy Hamil, Alex Aoyagi, Brett Duval, Amanda Baca, Francisco J Silva, Agnès Vallier, D Grant Jackson, Amparo Latorre, Robert B Weiss, Abdelaziz Heddi, Andrés Moya, Colin Dale
Affiliations
- PMID: 24407854
- PMCID: PMC3914690
- DOI: 10.1093/gbe/evt210
Genome degeneration and adaptation in a nascent stage of symbiosis
Kelly F Oakeson et al. Genome Biol Evol. 2014 Jan.
Abstract
Symbiotic associations between animals and microbes are ubiquitous in nature, with an estimated 15% of all insect species harboring intracellular bacterial symbionts. Most bacterial symbionts share many genomic features including small genomes, nucleotide composition bias, high coding density, and a paucity of mobile DNA, consistent with long-term host association. In this study, we focus on the early stages of genome degeneration in a recently derived insect-bacterial mutualistic intracellular association. We present the complete genome sequence and annotation of Sitophilus oryzae primary endosymbiont (SOPE). We also present the finished genome sequence and annotation of strain HS, a close free-living relative of SOPE and other insect symbionts of the Sodalis-allied clade, whose gene inventory is expected to closely resemble the putative ancestor of this group. Structural, functional, and evolutionary analyses indicate that SOPE has undergone extensive adaptation toward an insect-associated lifestyle in a very short time period. The genome of SOPE is large in size when compared with many ancient bacterial symbionts; however, almost half of the protein-coding genes in SOPE are pseudogenes. There is also evidence for relaxed selection on the remaining intact protein-coding genes. Comparative analyses of the whole-genome sequence of strain HS and SOPE highlight numerous genomic rearrangements, duplications, and deletions facilitated by a recent expansion of insertions sequence elements, some of which appear to have catalyzed adaptive changes. Functional metabolic predictions suggest that SOPE has lost the ability to synthesize several essential amino acids and vitamins. Analyses of the bacterial cell envelope and genes encoding secretion systems suggest that these structures and elements have become simplified in the transition to a mutualistic association.
Keywords: IS elements; comparative genomics; degenerative genome evolution; pseudogenes; recent symbiont.
Figures
Fig. 1.—
Microscopic images of SOPE. The main panel shows cells from a 5th instar bacteriome of SOPE stained with FM4-64 (red) and DAPI (blue). Rod-shaped bacteria are densely packed into the cytoplasm of the insect cells, whose nuclei display extensive DAPI staining. The inset panel shows isolated SOPE cells stained with DAPI, illustrating their filamentous morphology.
Fig. 2.—
Whole-genome sequence alignment of strain HS and SOPE. Bezier curves highlight regions of synteny shared between strain HS and SOPE. Uninterrupted blocks of synteny are rendered in the same hue. Matches occurring on the same DNA strand are rendered in the purple spectrum, whereas those occurring on different strands are rendered in the yellow spectrum. Thus, to maintain replicational symmetry, matches highlighted in purple should remain on the same replichore, whereas those highlighted in yellow should switch replichore. Sequences shared between the strain HS megaplasmid and SOPE chromosome are rendered in gray. The outer plots represent GC skew with positive skew depicted in red and negative skew depicted in blue. The strand-specific locations of KOPS (FtsK orienting polar sequences) sites are shown as tickmarks overlaid on the plot of GC skew. Inside the plots, the pink and blue tick marks correspond to the positions of phage and IS element sequences, respectively. These and any other forms of repetitive DNA were masked in the generation of the alignment. The location of the dif (terminus) sequence in strain HS is highlighted and intersects with the switch in GC skew, as expected.
Fig. 3.—
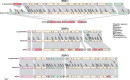
IS element dense regions in SOPE. Scalar illustration of two IS element dense regions in the SOPE chromosome. The top row corresponds to the region encompassing SOPEG_ps0144-SOPEG_0160, and the bottom row corresponds to SOPEG_2674-SOPEG_2683. IS element ORFs are colored according to their family (see key). ORFs are shaded in accordance with their positional synteny in comparison with a full length HS ortholog. The full spectrum of shading (5′–3′) is depicted in the key. ORFs disrupted by an IS element insertion are connected by curved lines. All non-IS element ORFs are labeled with their SOPE locus tags.
Fig. 4.—
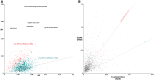
Plot of d_N_ versus d_S_ of orthologous genes in strain HS and SOPE. (A) Plot of d_N_ versus d_S_ of 1,601 orthologous genes in strain HS and SOPE. Each point depicted on the plot represents a single gene with the radius of each point proportional to gene size. Mean d_N_/d_S_ ratio is plotted as a gray dotted line. Genes with a d_N_/d_S_ ratio greater than 0.4 are annotated with their product. Genes with a d_N_/d_S_ ratio greater than 0.3 are depicted in red and those genes with d_N_/d_S_ ratios less than 0.3 are depicted in green. Mean ORF size was calculated for genes with d_N_/d_S_ greater than 0.3 (red) and d_N_/d_S_ less than 0.3 (green). (B) Plot of SOPE d_N_/d_S_ versus Sodalis glossinidius d_N_/d_S_ for 1,229 orthologous genes in strain HS, SOPE, and So. glossinidius. Each point on the plot represents a single orthologous gene.
Fig. 5.—
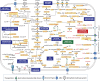
Overview of SOPE metabolism. The names in the yellow boxes indicate the genes predicted to be responsible for a given reaction. The generation of ATP is indicated. Abbreviations (besides the accepted symbols): CMP-KDO, CMP-3-deoxy-
d
-manno-octulosonate; DHF, dihydrofolate; ECA, enterobacterial common antigen; GlcNac-6P, N-acetyl glucosamine-6-phosphate; H/dH, nucleoside not G; DHAP, dihydroxyacetonephosphate, hydroxymethylpyrimidine; PEP, phospoenolpyruvate; PG, peptidoglycan; PRPP, phosphoribosyl pyrophosphate; SAM, S-adenosylmethionine; THF, tetrahydrofolate; UQ: ubiquinone.
Fig. 6.—
Comparison of TTSS gene clusters_._ Scalar illustration of the chromosomal regions encoding SSRs (SSR1–3) in Sodalis glossinidius (top), strain HS (middle), and SOPE (bottom). Genes are colored according to their predicted functions as secretion apparatus (tan), secreted effectors (green), chaperones (purple), transcriptional regulators (pink), hypothetical (white), or IS elements (brown). Pseudogenes are colored in red. Frameshifting indels and premature stops are shown as blue and red tick marks at their respective positions within the ORF. Pseudogenes without tick marks are inactivated by an IS element insertion within an ORF or by truncations of greater than 10% compared with the intact strain HS ortholog. Gene names are shown based on the strain HS annotation.
Similar articles
- Comparative genomics of insect-symbiotic bacteria: influence of host environment on microbial genome composition.
Rio RV, Lefevre C, Heddi A, Aksoy S. Rio RV, et al. Appl Environ Microbiol. 2003 Nov;69(11):6825-32. doi: 10.1128/AEM.69.11.6825-6832.2003. Appl Environ Microbiol. 2003. PMID: 14602646 Free PMC article. - Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host.
Toh H, Weiss BL, Perkin SA, Yamashita A, Oshima K, Hattori M, Aksoy S. Toh H, et al. Genome Res. 2006 Feb;16(2):149-56. doi: 10.1101/gr.4106106. Epub 2005 Dec 19. Genome Res. 2006. PMID: 16365377 Free PMC article. - Differential genome evolution between companion symbionts in an insect-bacterial symbiosis.
Bennett GM, McCutcheon JP, MacDonald BR, Romanovicz D, Moran NA. Bennett GM, et al. mBio. 2014 Sep 30;5(5):e01697-14. doi: 10.1128/mBio.01697-14. mBio. 2014. PMID: 25271287 Free PMC article. - Winding paths to simplicity: genome evolution in facultative insect symbionts.
Lo WS, Huang YY, Kuo CH. Lo WS, et al. FEMS Microbiol Rev. 2016 Nov 1;40(6):855-874. doi: 10.1093/femsre/fuw028. FEMS Microbiol Rev. 2016. PMID: 28204477 Free PMC article. Review. - The evolutionary history of symbiotic associations among bacteria and their animal hosts: a model.
Moya A, Gil R, Latorre A. Moya A, et al. Clin Microbiol Infect. 2009 Jan;15 Suppl 1:11-3. doi: 10.1111/j.1469-0691.2008.02689.x. Clin Microbiol Infect. 2009. PMID: 19220345 Review.
Cited by
- Flagella by numbers: comparative genomic analysis of the supernumerary flagellar systems among the Enterobacterales.
De Maayer P, Pillay T, Coutinho TA. De Maayer P, et al. BMC Genomics. 2020 Sep 29;21(1):670. doi: 10.1186/s12864-020-07085-w. BMC Genomics. 2020. PMID: 32993503 Free PMC article. - Mutualism breakdown by amplification of Wolbachia genes.
Chrostek E, Teixeira L. Chrostek E, et al. PLoS Biol. 2015 Feb 10;13(2):e1002065. doi: 10.1371/journal.pbio.1002065. eCollection 2015 Feb. PLoS Biol. 2015. PMID: 25668031 Free PMC article. - Relaxed selection underlies genome erosion in socially parasitic ant species.
Schrader L, Pan H, Bollazzi M, Schiøtt M, Larabee FJ, Bi X, Deng Y, Zhang G, Boomsma JJ, Rabeling C. Schrader L, et al. Nat Commun. 2021 May 18;12(1):2918. doi: 10.1038/s41467-021-23178-w. Nat Commun. 2021. PMID: 34006882 Free PMC article. - Antimicrobial peptides: Application informed by evolution.
Lazzaro BP, Zasloff M, Rolff J. Lazzaro BP, et al. Science. 2020 May 1;368(6490):eaau5480. doi: 10.1126/science.aau5480. Science. 2020. PMID: 32355003 Free PMC article. Review. - Enhanced Mutation Rate, Relaxed Selection, and the "Domino Effect" are associated with Gene Loss in Blattabacterium, A Cockroach Endosymbiont.
Kinjo Y, Lo N, Martín PV, Tokuda G, Pigolotti S, Bourguignon T. Kinjo Y, et al. Mol Biol Evol. 2021 Aug 23;38(9):3820-3831. doi: 10.1093/molbev/msab159. Mol Biol Evol. 2021. PMID: 34426845 Free PMC article.
References
- Akman L, et al. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat Genet. 2002;32:402–407. - PubMed
- Andersson JO, Andersson SGE. Genome degradation is an ongoing process in Rickettsia. Mol Biol Evol. 1999;16:1178–1191. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases