Regulation of TET protein stability by calpains - PubMed (original) (raw)
Regulation of TET protein stability by calpains
Yu Wang et al. Cell Rep. 2014.
Abstract
DNA methylation at the fifth position of cytosine (5mC) is an important epigenetic modification that affects chromatin structure and gene expression. Recent studies have established a critical function of the Ten-eleven translocation (Tet) family of proteins in regulating DNA methylation dynamics. Three Tet genes have been identified in mammals, and they all encode for proteins capable of oxidizing 5mC as part of the DNA demethylation process. Although regulation of Tet expression at the transcriptional level is well documented, how TET proteins are regulated at posttranslational level is poorly understood. In this study, we report that all three TET proteins are direct substrates of calpains, a family of calcium-dependent proteases. Specifically, calpain1 mediates TET1 and TET2 turnover in mouse ESCs, and calpain2 regulates TET3 level during differentiation. This study provides evidence that TET proteins are subject to calpain-mediated degradation.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no conflict of interests.
Figures
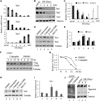
Figure 1. Regulation of TET protein levels by transcription and protein stability
(A) RT-qPCR analysis of Tet mRNA levels during mESC to NPC differentiation. While Tet1 and Tet2 levels decrease during differentiation, Tet3 level is significantly up-regulated. Data represent the mean of three independent experiments, and Tet levels in mESCs are set as 1. (B, C) Representative Western blot (B) and quantification of three repeats (C) demonstrate that TET protein levels generally follow mRNA levels during NPC differentiation. (D, E) Representative Western blot analysis of TET1 and TET2 levels in mESCs treated with chloroquine, calpeptin, Z-VAD-FAM and MG132 for 24h. Quantification of three independent experiments was shown in panel E. (F, G) Calpeptin increases the half-life of Flag-TET2 protein. Western blot (F) and quantification (G) of the Flag-TET2 levels in the presence or absence of calpeptin upon inhibition of protein translation by cycloheximide. (H, I) Representative Western blot analysis of TET3 in day 7 embryoid body (EB) treated with chloroquine, calpeptin, Z-VAD-FAM and MG132 for 24h, and the results were quantified in (I). (J) Calpain activity is detectable in mESCs and during their differentiation. Western blot analysis of mESC lysate with a spectrin antibody identified both full length (arrow) and cleaved spectrin (*), a marker for calpain activity. Spectrin cleavage is detectable during mESC differentiation (lanes 4, 5), and was prevented by calpeptin treatment (compare lane 1 and 2).
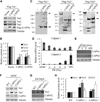
Figure 2. Tet proteins are direct substrates of calpain1 and calpain2
(A, B) Representative Western blot analysis (A) and quantification of three independent repeats (B) demonstrate that exogenously expressed TET protein levels can be reduced by co-expression of calpain1 or calpain2 in 293T cells. (C) Western blot analysis demonstrates that both calpain 1 and calpain 2 can cleave all three Tet proteins in vitro. Purified Flag-Tet1, Tet2 and Tet3 were incubated with buffer alone, or purified Flag-calpain1 or calpain2 at room temperature for 30min before Western blot analysis using Flag antibody. Cleaved products are indicated by (*), full length TET (arrow), calpains (arrow head). (D, E) RT-qPCR (D) and Western blot (E) analysis demonstrate that calpain1 and calpain2 are reversely expressed in mESCs and NPCs. Data represent the mean of three independent experiments, and value from mESC is normalized as 1. (F) Western blot analysis demonstrates that both TET1 and TET2 levels are increased in calpain1 knockout mESCs, while calpain2 knockout have little effect. Calpain1 and calpain2 knockout mESC were generated by CRISPR. (G) Western blot analysis of the TET3 levels in day 8 EB demonstrates calpain2 knockout increases TET3 levels, while the effect of calpain1 knockout is modest. (H) Quantification of three independent experiments (F and G), value in WT cells is set as 1.
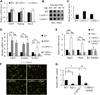
Figure 3. Effects of calpain-mediated TET cleavage on gene expression and NPC differentiation
(A) RT-qPCR analysis demonstrates that calpain1 or calpain2 knockout in mESCs does not affect pluripotent gene expression. Data represent the mean of three independent experiments, and value in WT mESC is set as 1. (B, C) Dot blot analysis (B) and densitometry quantification of three repeats (C) demonstrate that calpain1 knockout, but not calpain2, increased 5hmC levels in mESCs. (D) RT-qPCR analysis demonstrates that Tet1 knockdown in mESC enhances trophectoderm lineage genes (Cdx2 and Eomes) expression, and inhibits Lefty1. Knockout of calpain1 opposes this tendency, which is rescued by Tet1 knockdown. Data represent the mean of three independent experiments, and value in WT mESC is set as 1. (E) RT-qPCR analysis demonstrates that during differentiation to NPC (EB day 8), Tet3 knockout significantly reduces the expression of neuronal markers Ngn2 and Pax6, while calpain2 knockout enhanced their expression, which is reversed by Tet3 knockdown. In contrast, β3-tubulin expression is not affected by either Tet3 or calpain2. Value in WT EB is set as 1. (F) Immunostaining demonstrates generation of Nestin and Sox2 double positive NPCs. After EB disassociation and a 48h adherent culture, Nestin and Sox2 positive NPC were successfully generated from all WT and knockout cells. (G) TET3 and calpain2 have opposite effect on mESC differentiation to NPC. While Tet3−/− significantly reduced NPC generation, CAPN2−/− enhanced the differentiation efficiency, which is abolished by Tet3 knockdown. Number from WT cells is normalized to 1. P<0.05 (*), P<0.01 (**).
Similar articles
- Ten-Eleven Translocation 1 and 2 Confer Overlapping Transcriptional Programs for the Proliferation of Cultured Adult Neural Stem Cells.
Shimozaki K. Shimozaki K. Cell Mol Neurobiol. 2017 Aug;37(6):995-1008. doi: 10.1007/s10571-016-0432-6. Epub 2016 Oct 24. Cell Mol Neurobiol. 2017. PMID: 27778125 - Deletion of Tet proteins results in quantitative disparities during ESC differentiation partially attributable to alterations in gene expression.
Reimer M Jr, Pulakanti K, Shi L, Abel A, Liang M, Malarkannan S, Rao S. Reimer M Jr, et al. BMC Dev Biol. 2019 Jul 8;19(1):16. doi: 10.1186/s12861-019-0196-6. BMC Dev Biol. 2019. PMID: 31286885 Free PMC article. - Tet2- and Tet3- Mediated Cytosine Hydroxymethylation in Six2 Progenitor Cells in Mice Is Critical for Nephron Progenitor Differentiation and Nephron Endowment.
Liang X, Aranyi T, Zhou J, Guan Y, Hu H, Liu H, Susztak K. Liang X, et al. J Am Soc Nephrol. 2023 Apr 1;34(4):572-589. doi: 10.1681/ASN.2022040460. Epub 2022 Dec 15. J Am Soc Nephrol. 2023. PMID: 36522157 Free PMC article. - Mechanisms of TET protein-mediated DNA demethylation and its role in the regulation of mouse development.
Jia ZW, Gao SX, Zhang YC, Zhang XH. Jia ZW, et al. Yi Chuan. 2015 Jan;37(1):34-40. doi: 10.16288/j.yczz.2015.01.005. Yi Chuan. 2015. PMID: 25608811 Review. - Mechanisms that regulate the activities of TET proteins.
Joshi K, Liu S, Breslin S J P, Zhang J. Joshi K, et al. Cell Mol Life Sci. 2022 Jun 15;79(7):363. doi: 10.1007/s00018-022-04396-x. Cell Mol Life Sci. 2022. PMID: 35705880 Free PMC article. Review.
Cited by
- iPS cell generation-associated point mutations include many C > T substitutions via different cytosine modification mechanisms.
Araki R, Suga T, Hoki Y, Imadome K, Sunayama M, Kamimura S, Fujita M, Abe M. Araki R, et al. Nat Commun. 2024 Jun 11;15(1):4946. doi: 10.1038/s41467-024-49335-5. Nat Commun. 2024. PMID: 38862540 Free PMC article. - Denaturing purifications demonstrate that PRC2 and other widely reported chromatin proteins do not appear to bind directly to RNA in vivo.
Guo JK, Blanco MR, Walkup WG 4th, Bonesteele G, Urbinati CR, Banerjee AK, Chow A, Ettlin O, Strehle M, Peyda P, Amaya E, Trinh V, Guttman M. Guo JK, et al. Mol Cell. 2024 Apr 4;84(7):1271-1289.e12. doi: 10.1016/j.molcel.2024.01.026. Epub 2024 Feb 21. Mol Cell. 2024. PMID: 38387462 - The neuronal transcription factor MEIS2 is a calpain-2 protease target.
Müller T, Reichlmeir M, Hau AC, Wittig I, Schulte D. Müller T, et al. J Cell Sci. 2024 Feb 15;137(4):jcs261482. doi: 10.1242/jcs.261482. Epub 2024 Feb 28. J Cell Sci. 2024. PMID: 38305737 Free PMC article. - Increased iron uptake by splenic hematopoietic stem cells promotes TET2-dependent erythroid regeneration.
Tseng YJ, Kageyama Y, Murdaugh RL, Kitano A, Kim JH, Hoegenauer KA, Tiessen J, Smith MH, Uryu H, Takahashi K, Martin JF, Samee MAH, Nakada D. Tseng YJ, et al. Nat Commun. 2024 Jan 15;15(1):538. doi: 10.1038/s41467-024-44718-0. Nat Commun. 2024. PMID: 38225226 Free PMC article. - GID complex regulates the differentiation of neural stem cells by destabilizing TET2.
Xia M, Yan R, Wang W, Zhang M, Miao Z, Wan B, Xu X. Xia M, et al. Front Med. 2023 Dec;17(6):1204-1218. doi: 10.1007/s11684-023-1007-9. Epub 2023 Sep 14. Front Med. 2023. PMID: 37707676
References
- Bibel M, Richter J, Lacroix E, Barde YA. Generation of a defined and uniform population of CNS progenitors and neurons from mouse embryonic stem cells. Nature protocols. 2007;2:1034–1043. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 DK089565/DK/NIDDK NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 GM068804/GM/NIGMS NIH HHS/United States
- U01DK089565/DK/NIDDK NIH HHS/United States
- GM68804/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous